Abstract
Background:
Myelodysplastic syndromes (MDS) are highly heterogeneous regarding pathogenesis and clinical outcome. Traditionally, the revised international prognostic scoring system (IPSS-R)incorporating clinical features and cytogenetic abnormalities has been the standard for prognostication, while in recent years, recurrent somatic mutations are becoming increasingly important for this purpose. Additionally, gene expression profiling of coding genes and microRNAs has also emerged as a powerful tool to separate patients into distinct prognostic categories. Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without protein-coding capacity. lncRNAs not only participate in normal hematopoiesis, but perturbation of their expressions may contribute to development of acute leukemia. However, the impact of lncRNA expression profiling on the prognosis of MDS patients has not yet been explored to date.
Aims:
This study was aimed to evaluate lncRNAs expression profiling in MDS and to find out lncRNAs whose expression levels were associated with clinical outcomes. A scoring system was constructed to better risk-stratify MDS patients. We also tried to seek clues to the functionality of these lncRNAs.
Materials & Methods:
By using the Affymetrix Human Transcriptome Array 2.0 platform, we obtained the global expression profiles of 24120 lncRNAs in 176 adult patients with de novo MDS diagnosed according to the 2008 WHO classification. Through mathematical modeling, we identified six lncRNAs whose expression levels were significantly associated with overall survival (OS). We then constructed a risk scoring system with the weighted sum of each of these six lncRNAs, and evaluated the correlation of the scores with clinical features,cytogenetic abnormalities, gene mutations, andtreatment outcomesof the MDS patients.The reliability of our lncRNA scoring system was assured based on the coefficient of variance obtained from a permutation test, and further validated by using a five-fold cross-validation, in which 80% of the patients were used as the training samples to develop the scoring model, whose prediction performances were evaluated by using the rest of the 20% of the patients. Analysis of mutations in 21 genes was performed by conventional Sanger sequencing technique.
Results:
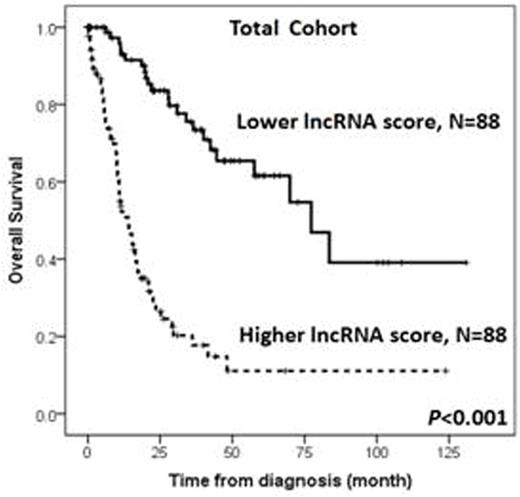
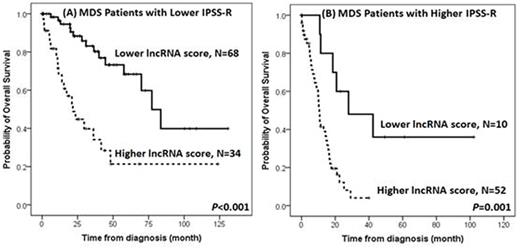
Higher lncRNA scores were positively associated with refractory anemia with excess of blasts (RAEB)-1 or RAEB-2 subtypes, complex cytogenetic changes, and IPSS-R high and very high risks, but inversely associated with RCUD, RARS, RCMD subtypes, a normal karyotype, and IPSS-R low risk. Patients with higher lncRNA scores had more frequent RUNX1, ASXL1, TP53, EZH2, SRSF2 and ZRSR2 mutations, shorter OS (median 14.0 months vs. 77.3 months, P<0.001, Figure 1), and higher five-year leukemic transformation rate (72.0% vs. 17.9%, P<0.001), compared with those with lower lncRNA scores. The predictive power of this 6-lncRNA model was validated by a five-fold cross validation procedure. In multivariable analyses, a higher lncRNA score remained an independent unfavorable risk factor for OS (RR 2.428, 95% CI 1.751-3.367, P<0.001) after adjusting for other clinical and molecular variables. Moreover, the survival difference between higher-score and lower-score patients remained statistically significant in both IPSS-R higher-risk (high and very high risks) and lower-risk (very low, low and intermediate risks) subgroups (Figure 2). We also correlated our lncRNA score with mRNA expression profiling, and identified that higher lncRNA scores were associated with pathways related to acute myeloid leukemia and hematopoiesis.
Conclusion:
Our integrated 6-lncRNA risk scoring system provides distinct insights into the clinical and biological implications of lncRNAs expression in de novo MDS patients. The scoring system represents an independent prognostic factor for survival and leukemic transformation. It may assist improving risk stratification of newly-diagnosed MDS patients. Further prospective trials are necessary to confirm the findings of our study.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal