Abstract
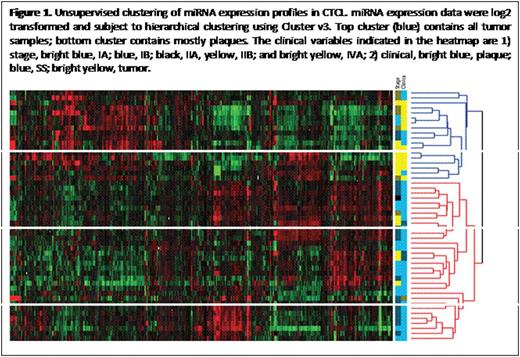
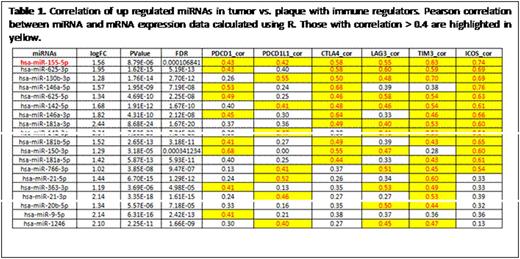
Cutaneous T cell lymphoma (CTCL) is thought to originate from clonally expanded, effector/central memory CD4+ T-cells in a background of chronic inflammation. Our recent investigations have provided evidence for T-cell exhaustion in CTCL with high levels of PD1 and other immune checkpoints (1. Gonzalez BR, Zain J, Rosen ST, Querfeld C. Current Opinion in Oncology. 2016; 28(1): 88-96. 2. Querfeld C, Curran SA, Leung S, Myskowski PL, Horwitz SM, Halpern AC, Young JW. Blood. 2014;124[21, abstract #1695). One potential mechanism by which immune checkpoint protein synthesis can be modulated is by the induction of miRNAs. A subset of miRNAs contributes to the pathogenesis and progression of mycosis fungoides/Sézary syndrome (MF/SS)), but no experimental research has evaluated the effects of miRNAs on immune checkpoints and T-cell exhaustion in CTCL. To address whether miRNA expression correlates with disease and is involved in regulating key immune checkpoint molecules linked to T cell exhaustion, we performed miRNA and gene expression profiling on 50 MF/SS patient samples using high-throughput sequencing, to achieve further insight into the molecular pathogenesis of CTCL disease. Total RNA, which includes miRNA, was extracted from CTCL (MF and SS) samples. Library preparation and high-throughput next generation small RNA sequencing were performed by the City of Hope Integrative Genomics Core. Reads were aligned to human genome assembly hg19, and miRNA expression levels were counted based on miRBase mature miRNA sequences. Differences in miRNA and gene expression between MF and SS patients were measured. Aberrantly expressed miRNAs specific for infiltrated plaques and tumor stage MF were compared. Our preliminary results have identified molecular signatures in CTCL that are linked to disease progression and key immunomodulatory molecules. Unsupervised cluster analysis of skin samples from patients using a heat map was performed (Fig 1). Patients were subdivided into 2 distinct clusters. The top cluster contains all tumor samples with few plaques, while the bottom cluster contains all patch/plaque samples. The clusters also correlated with clinical stage (top cluster = advanced stage ≥IIB; bottom cluster = early stage IA-IIA). Comparison analysis between the top and bottom cluster identified the 20 most significant up-regulated miRNAs that correlate with immune checkpoint (PD1, PD-L1, CTLA4, LAG3, TIM3, and ICOS) expression. For one, miR-146 showed the highest correlation (Table 1). The 20 most significant down-regulated miRNAs were also identified by class comparison analysis performed between top and bottom clusters and correlated with immune checkpoint expression. In conclusion, CTCL develops in a complex microenvironment. The expression of both miRNA and immune checkpoint proteins suggest patterns that provide insight into pathogenesis and potential therapeutic options.
Querfeld:Celgene: Consultancy, Research Funding; Actelion: Consultancy; Miragen: Consultancy. Zain:Seattle Genetics: Consultancy, Speakers Bureau; Celgene: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal