Abstract
High-throughput DNA sequencing of the adaptive immune receptor repertoire is a relatively new and fast growing technology used to study the immune response in health and disease. In B and T cell lymphoproliferative disorders, antigen receptor sequencing can be used to study clonal diversity and evolution of the disease in treatment free condition and in response to treatment. Furthermore, it can be used for the detection of minimal residual disease (MRD), providing information on the relationship between the presence and number of pre-treatment clone(s) and their relationship and responsibility for a subsequent relapse. The characteristics and quality of the data generated by high-throughput DNA sequencing of immune receptor signatures are the results of three major components: library preparation, sequencing platform, and software tools.
For both the library and software, there are no standard protocols and tools. Indeed, new approaches are continually being developed to accommodate new sequencing platform features and shortcomings, such as errors and read length restrictions. Two major technical challenges are: procuring an unbiased repertoire library that for B lymphocytes obtains and retains the full length IGHV-D-J along with (sub)isotype information, and resolving data to a single cell level, crucial for detection of MRD and rare clonal variants existing in the early phase of the disease, which might emerge and be involved in future relapse or progression.
We describe here a library preparation method for use with the Illumina MiSeq platform that results in an exhaustive full-length repertoire where virtually every B cell is sequenced, thereby maximizing the likelihood of identifying and quantifying the “real” IGHV-D-J repertoire of the sample analyzed. The method also allows the detection of very infrequent rearrangements and maintains IG sub-isotype information without compromising data quality. From 0.5 - 1 million human B cells can be sequenced in a single MiSeq 2x300 run with this approach. Key aspects of the technique are: 1) start from a well defined number of B lymphocytes 2) avoid V-gene specific PCR amplification and genetic material dilution in the pre-amplification phases 3) the specific depth of sequencing should depend on the starting B (or T) cell subset (i.e. na•ve, memory or plasma cell), and should be proportional to the number of starting cells. High quality sub-isotype information can be obtained with a second round of sequencing of shorter read length, e.g., with the Illumina 2x150 platform.
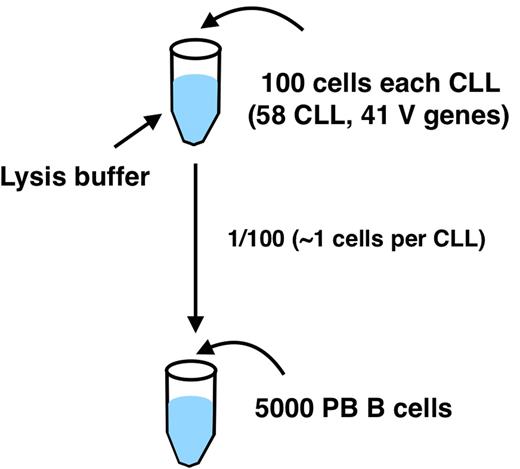
We used 58 different CLL clones with known IGH sequence mixed all together with polyclonal B cell from a donor PBMC (Figure 1). The mixed lysate is used to test the ability to detect the different clones.
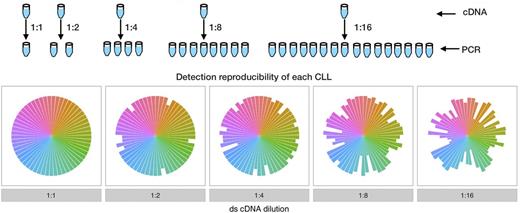
The following describes how the absence of genetic material dilution in the pre-amplification phases impact on the ability to obtain a comprehensive repertoire. These are crucial in MRD detection, since diluting the genetic material (RNA and/or cDNA) prior PCR amplification compromises the ability to accurately and consistently detect the clonal variants, reducing the de facto sensitivity and reproducibility of the analysis.
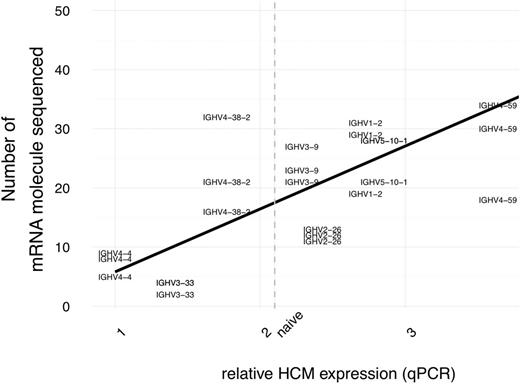
As a final example of the method's utility, we also demonstrate how different chronic lymphocytic leukemia clones present considerable variability in IG mRNA expression level that correlate with the number of unique mRNA molecule sequenced (Figure 3), which, if using a method with sub-optimal efficiency, could lead to a reduced clone-specific ability of detection by PCR based techniques.
Each dilution is performed in replicates. The cDNA is obtained from all the RNA extracted from the starting cells. Each slice represents a different CLL, and each slice size is the frequency for which it is detected. A comprehensive detection of each CLL is dependent to the absence of genetic material dilution.
Each dilution is performed in replicates. The cDNA is obtained from all the RNA extracted from the starting cells. Each slice represents a different CLL, and each slice size is the frequency for which it is detected. A comprehensive detection of each CLL is dependent to the absence of genetic material dilution.
qPCR IgH expression correlate with the number of unique mRNA molecule sequenced.
qPCR IgH expression correlate with the number of unique mRNA molecule sequenced.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal