Abstract
Leukemic clones within the same patient have unique sensitivities to drugs in vivo and may have unique capacities to engraft within xenograft models suggesting that there exist intra-patient functional clonal differences. However, methods to detect the functional differences assigned to genetically defined clones remain limited. Here, we describe a method to measure functional differences of clones within the same leukemia patient by coupling intracellular flow cytometry and next generation sequencing (NGS). We hypothesize that functionally distinct clones can be inferred by stimulating cells with cytokines, fluorescently staining with a relevant downstream marker of activation, and then isolating functionally distinct fractions by flow sorting. NGS can then digitally annotate somatic variants in sorted, functionally distinct samples that can be compared to the bulk leukemic cell population. To achieve this, we optimized a DNA isolation method that results in high-quality DNA despite originating from fixed/permeabilized cells. In this way, we annotate enrichment and de-enrichment of variants upon stimulation and infer the functional clonality of any given sample.
To provide proof-of-concept, we explored the well-established relationship between Chronic Myelmonocytic Leukemia (CMML) and GMCSF. CMML cells are uniquely hypersensitive to GMCSF and simultaneously have a well-established mutational composition that can be annotated by sequencing only 49 genes. This allowed us to leverage an amplicon-based enrichment panel that employs emulsion PCR to achieve reproducible library preparation with 5 ng of input DNA hence bypassing the need for whole genome amplification.
We tested our methodology by mixing two cell lines whose response to GMCSF was known and somatic variants had been annotated a priori. K562 (non-GMCSF responsive) and THP-1 (GMCSF responsive) leukemic cells were mixed at 1:1 and then stimulated with GMCSF. The resultant cells were stained with anti-pSTAT5 and cell sorting isolated positive and negative fractions. Unstimulated mixed cells and each positive and negative fraction was sequenced and variant allele fractions (VAF) was compared to unstimulated bulk (Figure 1A). To quantitate the change in VAF, we reasoned that summing the difference in VAF of unsorted/pSTAT5+ and unsorted/pSTAT5- populations would yield the most informative composite value as it would contain information about VAF difference for each mutation in both the GMCSF stimulated and GMCSF unstimulated fractions. Using this method, we found that 100% of THP1 and K562 variants were enriched in the pSTAT5+ and pSTAT5- fractions, respectively (Figure 1B). Receiver Operator Curves (ROC) demonstrated that the data functionally discriminated mixed THP-1 and K-562 cells with an AUC of 0.9868 (p<0.0001) (Figure1C). We additionally performed serial dilutions of THP-1 to K562 cell mixtures and demonstrated cell number dependent recovery of THP-1 variants upon stimulation with GMCSF.
Next we reasoned that somatic mutations should be uniquely augmented by GMCSF stimulation making it possible for somatic variant identification. To test this, we sequenced 8 CMML bone marrow mononuclear cells evaluated in the same way as above. We additionally sorted CD3+ cells as germline controls to identify somatic mutations. Using our 49-gene panel, we identified a total of 35 somatic variants and 160 germline variants. ROC curve analysis confirmed that our assay was capable of discriminating germline from somatic mutations with an AUC of 0.9388 (p<0.001).
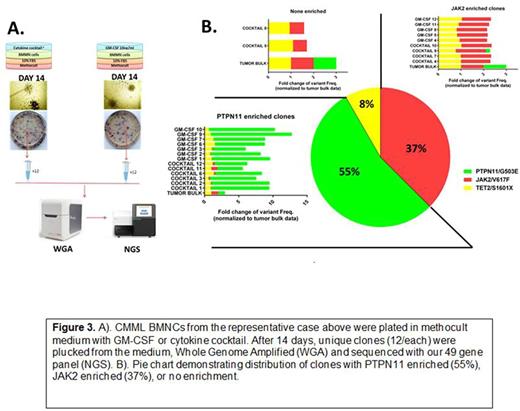
In a representative CMML case, we counterstained with CD33 to isolate myeloid progenitors and inferred a functionally distinct clone that was hypersensitive to GMCSF and uniquely carried a JAK2 and PTPN11 mutation that was obscured by bulk leukemia sequence (Figure 2). This inference was orthogonally validated by sequencing individual colonies from the same case grown under GMCSF conditions (Figure 3). Further validation is ongoing comparing mutations identified at engraftment of cells differentially xenografted in NSG and NSGS mice that differ only in their production of human cytokines. Taken together, we describe a novel method to annotate functional differences among mutationally-defined populations and identify functionally distinct clones in primary leukemia samples.
Komrokji:Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal