Abstract
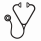
Background
Clonal hematopoiesis of indeterminate potential (CHIP) is an age-dependent genetic event occurring in as high as 10% of individuals over age 70. Although the clinical consequence of CHIP is not well understood, it is linked to an increased risk of primary hematologic malignancies and increased all-cause mortality. A recent study reported a higher prevalence (26%) of CHIP in patients with advanced cancers (Zehir A, ASCO abstract, 2016). However, the prevalence of CHIP in patients developing therapy-related myeloid neoplasms (T-MN) is unknown. We hypothesize that chemotherapy treated cancer patients with CHIP are at higher risk for T-MN development. To address this, we conducted a nested case-control study to compare the prevalence of CHIP in cancer patients who later developed T-MN compared to those that did not. We performed sequential targeted and whole exome sequencing of T-MN cases and described clonal evolution in cases for who paired CHIP and T-MN samples were available.
Methods
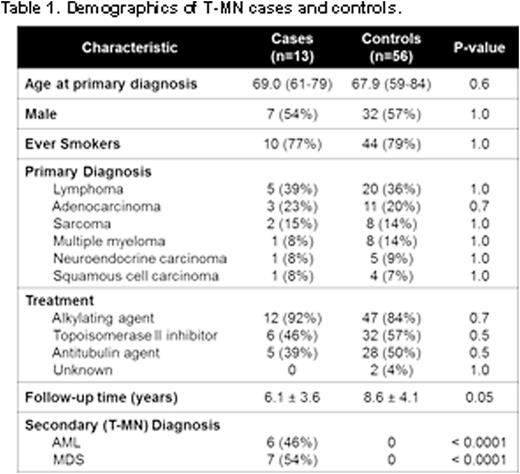
We identified all patients who consented to our institutional biobanking protocol with a primary cancer preceding T-MN and an age ≥ 70 years at the time of either cancer. Chart reviews were conducted to verify that only patients who received chemotherapy and provided a blood sample before development of T-MN were included. Controls were defined as cancer patients who did not develop a subsequent malignancy and were matched to cases 4:1 on gender, primary tumor diagnosis, age, smoking status (ever/never), chemotherapy drug class, and follow-up time.
DNA was isolated from peripheral blood (CHIP) or bone marrow samples (T-MN), and targeted amplicon based next-generation sequencing was performed to identify somatic mutations in genes associated with CHIP (49 genes, ThunderBolts Myeloid Panel, RainDance). Mesenchymal stem cells and CD3+ cells derived from the T-MN sample were used as independent germline controls to confirm somatic CHIP mutations. Whole exome sequencing was also performed (Agilent SureSelect XT Clinical Research Exome, Illumina NextSeq Sequencer) in paired CHIP and T-MN samples and clonal architecture was inferred using SciClone (Miller CA, PLoS Comput Biol, 2014). CHIP was defined in any sample that harbored a somatic mutation at an allele frequency of >3%.
Results
After screening 105,998 consented individuals from 2006 to 2016, a total of 13 patients (Figure 1a) were identified. The most common primary cancer in patients who developed T-MN was lymphoma, followed by adenocarcinoma (Table 1). The mean time to development of T-MN was 6.1 years and the mean time from the sampling date to the diagnosis of T-MN was 1.8 years. There were no statistical differences in the chemotherapy received by cases and controls (Table 1). The T-MN diagnosis was AML in 6 and MDS in 7 patients, and the majority (10/13) of patients harbored complex cytogenetics. T-MN patients had a poor overall survival (median 11 mo, Figure 1b). The prevalence of CHIP in cases and controls was higher (33%) than reported in individuals without cancer (10%), implying overlapping risk variables. Patients with T-MN had a significantly higher prevalence of CHIP prior to T-MN diagnosis when compared to matched controls (61.5% vs. 26.8%, p=0.02). The most commonly mutated gene in CHIP patients with T-MN was TP53 (36.4% vs. 5.4% in non-T-MN patients, p=0.02), while CHIP patients who did not develop T-MN more commonly harbored mutations in TET2 (35%) (Fig 1c). There was no difference in the mean allele frequencies (MAF) of CHIP mutations in cases vs. controls (9.4% vs. 14.3%, p=0.27). There was also no trend in the number or MAF of CHIP mutations observed and time to T-MN. In the majority of cases (83.3%), CHIP MAF expanded at the time of T-MN, however, a subset of paired samples (16.7%) harbored CHIP mutations that retracted giving way to expansion of a distinct mutant clone (Fig 1d).
Conclusions
Chip often precedes development of T-MN in chemotherapy-treated cancer patients. The distribution of CHIP-related gene mutations differs between individuals with T-MN vs. those without suggesting that there may be mutation-specific differences in T-MN risk. Further, although most CHIP mutations represented the founding leukemic mutation based on allelic frequency, some CHIP mutations retracted at the time of T-MN. These studies support the use of CHIP to inform individualized T-MN risk across cancer.
Komrokji:Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Speakers Bureau. Padron:Incyte: Research Funding; CTI: Research Funding; KALOBIOS: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal