Abstract
As relapses in precursor T-ALL are frequently refractory to treatment, patients at high risk of relapse need to be identified and treated with intensified or novel regimens. Because minimal residual disease (MRD) assessment identifies patients at high risk only after induction treatment has been completed, there is an urgent need for genetic markers that allow for early risk stratification. We aimed at identifying such markers by analyzing a cohort of patients treated by ALL BFM 2000 and AIEOP-BFM ALL 2009 protocols for CNAs in both established leukemia driver genes and in a set of nine novel candidate genes that had been found to be frequently affected by gene panel sequencing.
We analyzed bone marrow DNA of a large cohort of children with T-ALL for copy number alterations (CNA) by multiplex ligation dependent probe amplification (MLPA) using the commercially available probe set P383 (MRC Holland) and a custom made probe set targeting additional 9 genes (STAT5B, CNOT3, CNOT6, CTCF, ABCA5, PDGFRB, MYC, PTK2B, AKT1).
We correlated the occurrence of these CNAs with risk of relapse and found that none of the CNAs listed in tables 1 and 2 as a single alteration predicted the risk of relapse in any risk group. By contrast, the combination of CNAs can define risk signatures. Specifically, we found the frequently linked deletions of CDKN2A and MLLT3 on chromosome 9p21.3 to have distinct effects: While biallelic CDKN2A-deletions increase the risk of relapse (Hazard ratio 2.1, p(χ)=0.049), this effect is counteracted by deletions of MLLT3, which reduce the risk of relapse (hazard ratio 0.35, p(χ)=0.031). Consequently, a combination of CDKN2A and MLLT3 deletions, together with activating mutations of NOTCH1, defines three risk groups in the German cohort of 209 patients treated in the intermediate and high risk groups of the ALL BFM 2000 protocol: Patients that carry both a NOTCH1 mutation and an MLLT3 deletion, but not a biallelic CDKN2A deletion, have a 5 year cumulative incidence of relapse (CIR) of 3% (1/29), whereas patients with biallelic CDKN2A deletion without MLLT3 deletion have a 5 year CIR of 26% (20/78; p=0.0057). All remaining patients have a 5 year CIR of 10 % (10/102). Notably, this risk signature might be highly dependent on the treatment context, because it predicted risk neither in 119 AIEOP-BFM 2009 patients treated in Czech Republic and Germany nor in 80 ALL BFM 2000 patients treated in Austria.
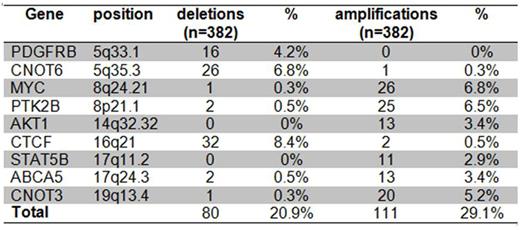
We next considered the MRD-defined high-risk (HR) group of the BFM 2000 and AIEOP-BFM 2009 protocols consisting of 37 patients. At least one of the 9 CNAs listed in table 2 was detected in 64 of 376 patients of whom a complete set of clinical data were available and in 11 of 37 HR-patients (p=0.04). The presence of any of these 9 CNAs splits the HR group in a smaller "ultra-high risk group" with a 5 year CIR of 82% (8/11) and a larger group of patients with a 5 year CIR of only 24% (6/26) (p(Gray)=0.005). Neither was there an effect in MRD-MR patients nor in patients who were HR defined by prednisone-poor-response only. It must be noted that the deletions and amplifications shown in table 2 are part of large CNAs. Therefore, the indicated genes do not necessarily define risk per se but may represent markers for other changes with functional relevance or may indicate a more general genomic instability.
We conclude that complementing MRD risk parameters with an analysis of common CNAs refines the current risk stratification and may identify an ultra-high risk group of children with T-ALL, who are candidates for experimental treatment even in primary disease.
Frequency of known and functional CNAs detected by P383 in BFM 2000 and BFM 2009 T-ALL patients
Frequency of known and functional CNAs detected by P383 in BFM 2000 and BFM 2009 T-ALL patients
Frequency of CNA defined by novel candidate genes in BFM 2000 and 2009 patients
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal