Abstract
Background: Prognostication in MF has historically been based on age, symptoms and hematologic features at presentation. Newer models include cytogenetic and genomic data given their independent prognostic value. In 2014, two prognostic models incorporating gene mutation data were proposed: a "mutation enhanced" IPSS (MIPSS) and a genetics-based prognostic scoring system (GPSS) model; the former using mutation data to enrich standard prognostic inputs, while the latter eschewed classic prognostic inputs for a score comprised solely of cytogenetics, mutation data, and age (Vannucchi, Blood, 2014 and Tefferi, Blood, 2014). In this study, we aimed to validate these risk models as well as propose age-independent models to better capture disease-specific prognosis.
Methods: This was a single institution, retrospective study of confirmed MF patients seen between 2/2001 and 6/2016. Definitions of primary myelofibrosis (PMF), post-essential thrombocythemia myelofibrosis (post-ET MF) and post-polycythemia vera myelofibrosis (post-PV MF) were according to World Health Organization 2016 criteria and the International Working Group for Myeloproliferative Neoplasms, Research and Treatment, respectively. Mutation and cytogenetic data were assessed from review of electronic medical record. Prognostic scores were calculated on first presentation to our facility. Survival curves were generated using Kaplan-Meier method and compared using the log-rank test.
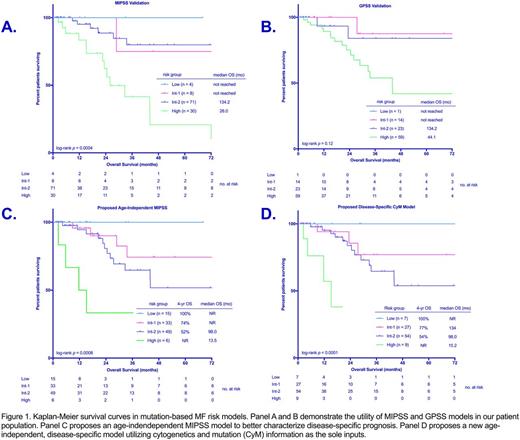
Results: We identified 103 and 97 patients with available data to calculate MIPSS and GPSS scores respectively. The cohorts were 60% male, with median age 72 years at presentation. The majority of patients had a diagnosis of PMF (80%), with a small component of post-ET MF (13%) and post-PV MF (7%). Assigning scores based on MIPSS, 4% of patients were low-risk (LR), 8% intermediate-1 (IR1), 60% intermediate-2 (IR2), and 28% high-risk (HR). The model successfully differentiated between IR2 and HR with median overall survival (OS) 134 and 26 months, respectively (p = 0.0001) (figure 1A). No significant difference in OS was seen between LR/IR1 groups and IR2 (p = 0.58). Using GPSS, 1% of patients were classified as LR, 13% IR1, 25% IR2, and 61% HR. GPSS performed well in delineating between HR and intermediate risk (combined IR1 and IR2) (median OS 44 vs 134 mo, p = 0.02); however it did not distinguish HR from IR2 (p = 0.09) nor between IR1 and IR2 (p = 0.71) (figure 1B).
To more specifically assess disease-specific prognosis and better divide risk scores among our patient population, new models were created. Given that age is a negative risk factor for OS independent of disease state, this input was removed. First, an age-independent MIPSS score was created with four risk groups defined: LR (score 0-0.5), IR1 (score 1-1.5), IR2 (2-3), and HR (score 3.5+). Fifteen percent of patients were classified as LR, 32% IR1, 48% IR2, and 6% HR. This system distinguished significantly between IR2 and HR (median OS 98.0 vs 13.5 mo, p = 0.006), while providing improved differentiation between IR1 and IR2 risk groups (median OS not reached vs. 98 months, p = 0.09) (figure 1C).
Next, we aimed to create a truly disease-specific model based on the GPSS. Age was omitted and the impact of SRSF2 mutation was enhanced from 1 point to 1.5 points given its greater weighted impact on OS compared to ASXL1 mutation in our database. Values for other inputs remained consistent with GPSS. Using cytogenetic information (CY) and mutation data (M) as the sole inputs a CYM score was created. This yielded four risk groups defined as LR (0 points), IR1 (1-2.5 points), IR2 (3-5 points) and HR (5.5+ points). Seven percent of patients were classified as LR, 28% IR1, 56% IR2, and 9% HR. This model differentiated between IR2 disease and HR disease effectively (median OS 98 vs 15.2 mo., p < 0.0001). No significant difference was observed between IR1 and IR2 disease (figure 1D).
Conclusion: Using previously proposed molecular-based models, we were able to validate their utility in the setting of advanced disease; however, insufficient capture of lower risk categories impaired its validity in that setting. We propose age-independent models to better characterize disease-specific risk in an effort to identify patients who many benefit from allogeneic stem cell transplant and/or clinical trial referral. These models merit testing in larger cohorts with longer follow-up to determine their clinical validity.
Lancet:Celgene: Consultancy, Research Funding; Amgen: Consultancy; Seattle Genetics: Consultancy; Quantum First: Consultancy; Biopath Holdings: Consultancy; Baxalta: Consultancy; Pfizer: Research Funding; Kalo Bios: Consultancy; Karyopharm: Consultancy; Jazz Pharmaceuticals: Consultancy; ERYtech: Consultancy; Boehringer-Ingelheim: Consultancy; Novartis: Consultancy. Sweet:Pfizer: Speakers Bureau; Novartis: Consultancy, Speakers Bureau; Ariad: Consultancy, Speakers Bureau; Incyte Corporation: Research Funding; Karyopharm: Honoraria, Research Funding. Komrokji:Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal