Abstract
Background: Advances in detailing the interaction between tumor cells and host immune system has led to the potential of targeted immunotherapy. One target is the programmed death 1 (PD-1) pathway, which cancer cells up-regulate to evade T-cell-mediated immune response. Blockade of PD-L1 or PD-L2 has shown efficacy in solid tumors, as well as classic Hodgkin's lymphoma (cHL) and primary mediastinal b-cell lymphoma (PMBCL). In cHL and PMBCL, amplification or rearrangement of chromosome 9p24.1 is implicated in their pathogenesis. The genes PDL1 and PDL2 are located on 9p24.1, along with JAK2, which further induces PDL1 and PDL2. Targeted inhibitors of JAK2 are utilized in some myeloid malignancies (MM), mostly as gain of function point mutations. The aim of this study is to determine the prevalence of and classify 9p24.1abnormalities across subtypes of MM to guide future studies for targeted therapy.
Methods: A service of the National Cancer Institute, theMitelman Database of Chromosome Aberrations and Gene Fusions in Cancer database is manually updated from the literature and organized into three sub-databases: cases of chromosomal aberrations, molecular biology and clinical associations, and references. We searched for the 9p24 breakpoint within hematologic malignancies with no other search limits and subsequently divided the cases into myeloid and lymphoid subtypes. The incidence of rearrangements and additions in chromosome 9p24 for myeloid subtypes were determined. Those with either incidence >0.5% ort(8;9)(p21-23;p23-24) translocations are reported.
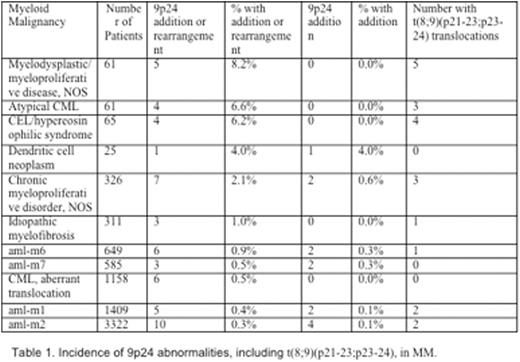
Results: 9p24 alterations occurred in 92 of 25,284 cases (<0.4%) of MM across 24 subtypes, with 22 (<0.1%) of these additions. Of subtypes with at least 100 cases, 9p24 alterations in chronic myeloproliferative disorder, NOS (7/326, 2.1%) and idiopathic myelofibrosis (3/311, 1.0%) were the most common. Chronic myeloid leukemia (CML),t(9;22),myelodysplastic syndrome and acute myeloid leukemia (aml) had <1% incidence of 9p24 rearrangements and additions. Myelodysplastic/myeloproliferative disease, NOS (5/61, 8.2%), atypical CML (4/61, 6.6%) and chronic eosinophilic leukemia (CEL)/hypereosinophilic syndrome (4/65, 6.2%) contained the most frequent 9p24 alterations. Notably,t(8;9)(p21-23;p23-24) translocations accounted for 21/92 (22.8%) MM 9p24 alterations and were seen in 8 subtypes:myelodysplastic/myeloproliferative disease, NOS (5 cases), CEL/hypereosinophilic syndrome (4), atypical CML (3), chronic myeloproliferative disorder, NOS (3), aml-m1 (2), aml-m2 (2), aml-m6 (1), and idiopathic myelofibrosis (1). One patient with aml-1 and one with chronic myeloproliferative disorder was found with t(9;22)(p24;q11).
Conclusions: Despite a low incidence of 9p24 rearrangements and additions across all MM within the Mitelman database, t(8;9)(p21-23;p23-24) translocations were seen in 8 MM subtypes and accounted for 22.8% of observed alterations. This translocation results in the fusion of PCM1-JAK2, leading to activation of Janus Kinase 2. Other than two patients with t(9;22)(p24;q11), harboring fusion of BCR-JAK2,no other rearrangements involving JAK2 were identified.While the use of JAK2 inhibitorruxolitinib has been described in 3 patients with t(8;9)(p21-23;p23-24), 2 with chronic eosinophilic leukemia and 1 with myeloproliferative disease, other MM subtypes with this translocation have not been evaluated. Further, no study has assessed such cases with FISH and other testing to streamline potential forPD-L1 or PD-L2 inhibitors. This data supports the utility of theMitelman database to identify alterations that can be further evaluated for targeted therapy and clinical trials.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal