Abstract
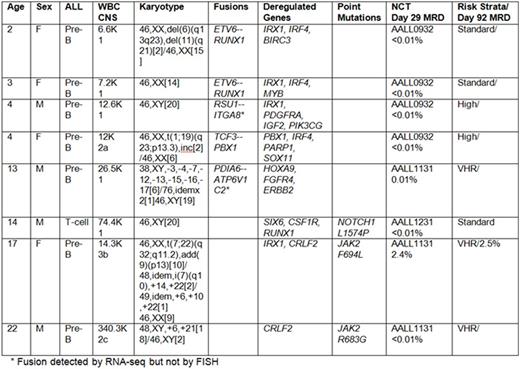
The biology and treatment of acute lymphoblastic leukemia (ALL) continues to attract attention as an area for which targeted therapies offer great promise. However, the integration of targeted therapies remains a challenge, both in identifying targetable genomic alterations, and the practical challenge of how best to integrate targeted therapies into clinical practice. To address these challenges in children and young adults, we developed a feasibility study to assess the prevalence of targetable genomic alterations in our population. We hypothesized that within our patient population, a spectrum of novel genomic alterations could be identified and characterized early in the treatment course. In cases where actionable alterations were identified, we further hypothesized that inclusion of novel therapies into standardized regimens might prevent relapse. In accordance with the Declaration of Helsinki, we obtained written, informed consent from eligible study subjects to perform genome sequencing in newly diagnosed patients. We enrolled 8 participants; 4 males and 4 females, ranging in age from 2 to 22 years (Table). Samples underwent deep sequencing of more than 400 cancer relevant genes (Ion Ampliseq Comprehensive Cancer Panel, Thermo Fisher) and unbiased RNA sequencing. In accordance with our institutional practices, all 8 participants underwent standardized diagnostic testing to ascertain CNS status, cytogenetics and induction responses before assignment/randomization onto a Children's Oncology Group (COG)-sponsored therapeutic study. Seven subjects had B-ALL and one had T-ALL. Genomic and molecular aberrations were identified in all patients. Using NCI risk factors, karyotypic features and Day 29 response metrics, 2 were treated with standard-risk B-ALL therapies (COG AALL0932), 1 with T-ALL (COG AALL0434) and 5 with high-risk B-ALL (COG AALL1131). Two patients started with standard-risk therapy, but were escalated to high-risk treatment based on elevated day 8 peripheral blood MRD. Genomic testing revealed that two patients harbored mutations in JAK2; one with a known mutation site and one with a novel F694L mutation. While all subjects entered into morphologic remission, our 17-year old patient with a novel JAK2 F694L mutation had high persistent MRD, suggesting she was at risk for an on-therapy relapse. With the novel JAK2 mutation confirmed in a CLIA setting, we integrated ruxolitinib into her post-induction therapy. Over the course of three month's time, her MRD diminished from 10% to 0.007%. Following a related, allogeneic bone marrow transplant, she remains in CR1. The second patient with a JAK2 R683G mutation relapsed after 20 months amid concerns of non-compliance with his maintenance therapy. Genomic testing on the relapse clone identified the same JAK2 R683G mutation that was identified at diagnosis. Our results show that prospective testing and therapy modifications can be achieved in real-time, allowing patients who might have suffered relapse to achieve adequate disease control for high-risk acute lymphoblastic leukemia.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal