Abstract
The role of metabolic dysfunction in the initiation and progression of AML is still to be determined. Next-generation sequencing studies in AML have identified somatic mutations in mitochondrially encoded subunits of the mitochondrial respiratory chain (MRC; N Engl J Med, 2013; 368:2059). Recurrent somatic mutations in nuclear MRC genes are yet to be reported, but recurrent founding mutations occur in nuclear genes encoding the TCA cycle enzymes Isocitrate Dehydrogenase 1 and 2 (IDH1, IDH2). IDH1 and IDH2 mutant AML samples display sensitivity to BCL-2 inhibition (Nat Med, 2015; 21:178), and reduced proliferative capacity has been observed for IDH1 mutant samples following glutaminase inhibition (Exp Hematol, 2014; 42:247). In glioma, IDH2 mutant tumors show enrichment of oxidative phosphorylation (OXPHOS) related gene sets (J Exp Clin Cancer Res, 2016;35:86). Additionally, germline succinate dehydrogenase (MRC Complex-II) mutations have been associated with hereditary paragangliomas (Science, 2000; 287:848). We therefore investigated the nature and frequency of rare germline and somatic mutations affecting nuclear MRC genes in an adult AML cohort, and profiled the metabolic phenotypes of primary BM AML samples.
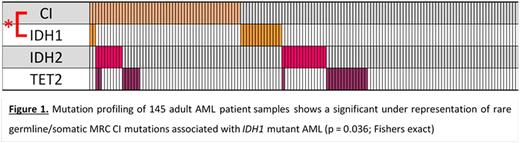
Whole-exome sequencing was performed on 145 diagnosis (Dx) adult BM AML samples. Variants were filtered against dbSNP137, 1000 genome and NHLBI-ESP with a focus on rare germline/somatic variants with a minor allele frequency < 0.005. A total of 62 variants affecting the 39 nuclear MRC Complex-I (CI) genes were found in 52 samples. Across the MRC (85 nuclear genes), we identified 140 variants in 95 samples. Confirmation of the germline/somatic status of identified variants is ongoing, with 20 / 21 variants tested being germline in origin. Case-control burden analysis (Am J Hum Genet, 2013; 92:841) was performed to compare variants in the AML cohort with those in an ethnically matched healthy control cohort (n = 329; PLoS Genet, 2011; 7:e1001372). This showed enrichment in the AML cases for variants across all 5 MRC complexes (p = 0.04; Burden analysis, Bonferroni adjusted), and for variants affecting the β-complex of CI (14 genes; p = 0.01; Burden analysis, Bonferroni adjusted). We further investigated the association of CI variants with genes commonly mutated in AML. This revealed a significant under-representation of IDH1 mutations in AML with CI variants (p = 0.036; Fisher's exact; Figure 1).
Additionally, for a panel of AML samples (n = 117), we used the BioMark (Fluidigm) qRT-PCR platform to profile the expression of all nuclear CI genes and a panel of AML oncogenes and tumor suppressors. This revealed a significant negative correlation (p < 0.05; Pearson's correlation, Bonferroni adjusted) between the expression of all nuclear CI genes and that of IDH2, which is independent of underlying mutation or genetic alterations, and which is not observed for IDH1 expression. Finally, Seahorse metabolic profiling showed that IDH1 mutant samples were found to be utilizing their glycolytic pathway at maximal capacity with no glycolytic reserve while simultaneouslydisplaying significantly reduced glycolytic rates (p< 0.05; T-test, Bonferroni adjusted) when compared to IDH2 mutant AML samples, AML samples lacking recurrent AML mutations (WT-AML), and normal bone marrow (NBM) controls. Both IDH1 and IDH2 mutant samples showed significantly deficient OXPHOS reserve capacity when compared to NBM and WT-AML samples (p < 0.05; T-test, Bonferroni adjusted). Ongoing investigations will determine the metabolic phenotype of CI mutant samples.
In summary, we observed significant enrichment of rare variants affecting the MRC and CI genes in an adult AML cohort compared to healthy controls, highlighting a potential role for altered cellular energetics in AML pre-disposition. The significant under-representation of somatic IDH1 mutations in AML with rare germline/somatic CI mutations raises the possibility that these CI variants induce functional consequences that mimic those associated with somatic IDH1 mutation. The distinct glycolytic profiles of IDH1 and IDH2 mutant samples, and the selective negative correlation of CI gene expression with IDH2 mRNA expression, suggests that distinct metabolic phenotypes maybe associated with perturbations to IDH1 and IDH2. Ongoing investigations will compare the metabolic phenotype of samples with CI variants to that of IDH1 and IDH2 mutant samples.
Gill:Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal