Abstract
Introduction
Advances in genetic sequencing have shed light on the biological underpinnings of AML, however, the vast majority of previous work has focused on young patients, with little evidence-based data in the elderly population. Older age at diagnosis is a well-known poor prognostic factor, however prognostication within the older age group itself remains a challenge making therapeutic decision making particularly difficult. In this pilot project we studied the transcriptomes of short and long-lived elderly AML patients to gain insight into the potential molecular differences and signaling pathways that may help better prognosticate patients within this unique population.
Methods
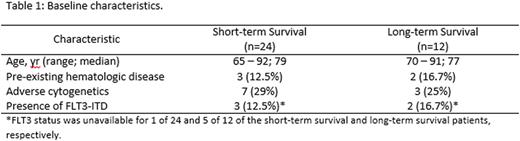
Elderly patients (age > 65 and not fit for induction chemotherapy) with newly diagnosed AML (excluding APL) between 2011 - 2015, inclusive, with an available diagnostic bone marrow biopsy sample were considered for inclusion in this retrospective analysis. Patients were divided into two groups: long-term survivors (survival ≥ 6 months) and short-term survivors (survival < 2 months). RNA sequencing was performed on 12 patients in the long-term survivors group and 24 patients in the short-term survivors group.
RNA sequencing was conducted using the Illumina platform (Illumina NextSeq 500) and data analysis was performed with TopHat and Cufflinks software.
Results
Baseline clinical characteristics were similar between the short-term and long-term survival groups as shown in Table 1.
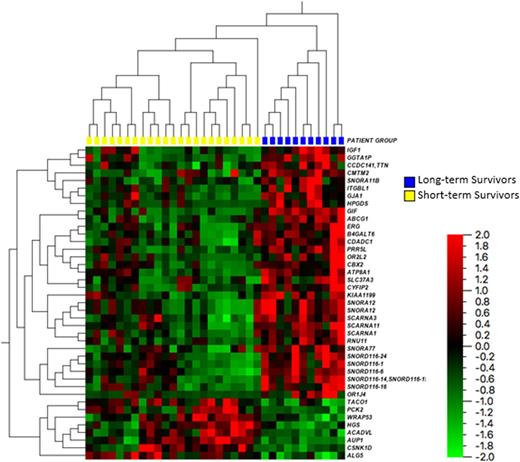
RNA sequencing revealed 41 genes with statistically significant (p-values < 0.001 and false-discovery q-values < 0.001) differential expression between the long-term and short-term survival groups. See Figure 1 for a heat map reflecting the gene expression values between groups. Of these 41 genes several are known to be involved in key cellular functions and signaling pathways including RNA post-transcription regulation, apoptosis, p53 regulation, and the mTOR pathway. However, only a few have previously been studied in AML (e.g. ERG, PCK2, and ABCG1) and none have been examined in the context of elderly AML patients.
Twelve of the 41 differentially expressed genes were small nucleolar RNAs (snoRNAs), a class of regulatory RNAs involved in post-transcriptional modification of ribosomal RNA. These were found to be down regulated in the short-term survivors compared to the long-term survivors (p-value range 0.00005 - 0.001). This is a novel finding. Although recent studies have found differences in snoRNA expression in AML and ALL compared to healthy donors there are no published studies examining the role of snoRNA in the prognosis of AML.
CYFIP2 is involved in caspase activation and cellular apoptosis and was found to be relatively under expressed in the short-term survivors group (p-value 0.008). WRAP53 plays an important role in the regulation of p53 expression and was found to be under-expressed in the long-term survivors. PRR5L is associated with mTORC2 and was found to be relatively over-expressed in the long-term survivors (p-value 0.00002). Due to the small sample sizes of this pilot project multivariate analysis was not conducted.
In addition to the individual genes, these results highlight differences in several pathways, namely the mTOR, and p53 tumor suppressor/caspase apoptotic pathways, which may be associated with prognosis for elderly patients with newly diagnosed AML and deserve further investigation. The finding of down regulation of numerous snoRNAs in elderly patients with poor outcome also warrants further detailed study with larger sample sizes to fully elucidate their potential prognostic value.
Figure 1: Heat Map highlighting the differential gene expressions from RNA sequencing for long-term compared to short-term survivors.
Conclusion
We have identified distinctly different gene expression profiles in elderly AML patients with long-term compared to short-term survival. These differentially expressed genes provide biologic insight into AML in the elderly as well as highlight candidate pathways and cellular mechanisms on which to base future detailed study to enable accurate prognostication and improved therapeutic decision making in this understudied population.
Owen:Roche: Honoraria, Research Funding; Janssen: Honoraria; Lundbeck: Honoraria, Research Funding; Abbvie: Honoraria; Novartis: Honoraria; Gilead: Honoraria, Research Funding; Pharmacyclics: Research Funding; Celgene: Honoraria, Research Funding. Geddes:Celgene: Other: Advisory Board, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal