Abstract
The discovery that mRNA splicing factors are frequently mutated in clonal blood disorders represented one of the most unexpected findings from cancer genome sequencing projects. While initial functional studies of these mutations have suggested that alterations in constitutive and alternative splicing may directly contribute to malignant hematopoiesis, they also highlighted limitations in our knowledge of how splicing factors regulate differential splicing and usage of mRNA isoforms in normal hematopoietic development. Interestingly, the splicing factor SRSF2, a member of the serine/arginine-rich (SR) protein family that regulates alternative splicing in a tissue-specific manner, is recurrently mutated only in myeloid leukemias, whereas other splicing factors are also often mutated in lymphoid leukemias and epithelial cancers. These data suggest a specific role for SRSF2 in hematopoiesis and/or myelopoiesis and previous studies have shown that Srsf2 is required for T-lymphopoiesis and hematopoietic stem and progenitor cell (HSPC) function. At the same time, SRSF2s functions in regulating splicing have been suggested to be at least partially overlapping with those of SRSF1, the founding member of the SR protein family. We therefore set out to understand the role and function of SRSF1 in normal fetal and adult hematopoiesis and to directly compare SRSF1s role in normal hematopoiesis to that of SRSF2.
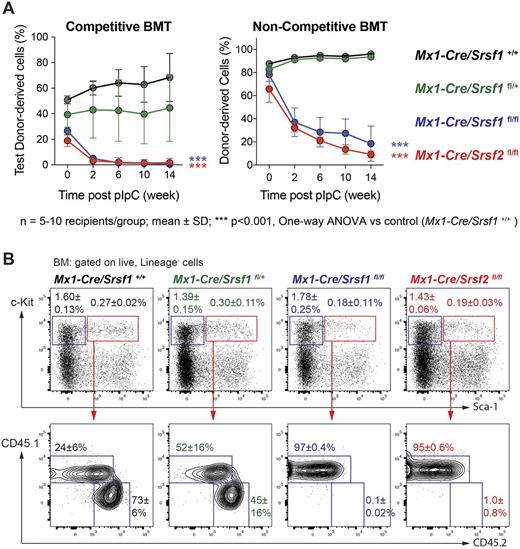
To understand the role of Srsf1 in adult hematopoiesis, we utilized mice for conditional deletion of 1 or 2 copies of Srsf1 or Srsf2 and performed non-competitive and competitive bone marrow transplantation (BMT) assays. BM from adult Mx1-Cre/Srsf1+/+, Mx1-Cre/Srsf1fl/+, Mx1-Cre/Srsf1fl/fl and Mx1-Cre/Srsf2fl/fl mice (together with the same number of wildtype CD45.1 marrow in competitive BMT) were transplanted into lethally irradiated CD45.1 recipients, followed by polyinosinic:polycytidylic acid (pIpC) administration 4 weeks after reconstitution to induce Srsf1 deletion. In both competitive and non-competitive BMT, peripheral blood (PB) chimerism showed that while heterozygous deletion of Srsf1 did not affect PB chimerism, homozygous deletion of Srsf1 resulted in compromised multi-lineage reconstitution similar to homozygous Srsf2 deletion (Figure 1A). Analysis of hematopoietic organs 20 weeks post BMT revealed no contribution to hematopoiesis by Srsf1 or Srsf2 homozygous knockout cells. More specifically, the contribution of HSPC-enriched Lineage- Sca-1+ c-Kit+ (LSK) and myeloid progenitor-enriched Lineage- Sca-1- c-Kit+ (LK) fractions were significantly reduced in mice transplanted with Srsf1- and Srsf2-deficient BM (Figure 1B). These observations identify an absolute requirement for both splicing factors in HSPC function in adult mice.
To determine if Srsf1 or Srsf2 are required for fetal hematopoiesis, we crossed Vav-CreTg/Srsf1fl/+ to Srsf1fl/fl mice and Vav-CreTg/Srsf2fl/+ to Srsf2fl/fl mice followed by HSPC analysis at embryonic day 13~14. At this time point, both Vav-CreTg/Srsf1fl/fl and Vav-CreTg/Srsf2fl/fl embryos were found at the approximate expected Mendelian ratio (p=0.77 for Srsf1 crosses and p=0.22 for Srsf2 crosses; Fishers exact test). Interestingly, the numbers of HSPCs and erythroid progenitors (determined by flow cytometry using Ter119 and CD71 antibodies) were similar in Vav-CreTg/Srsf1fl/fl fetal livers compared to control embryos, while Vav-CreTg/Srsf2fl/fl fetal livers had drastically reduced fetal liver cellularity, HSPC, and erythroid progenitors relative to control embryos. However, both Vav-CreTg/Srsf1fl/fl and Vav-CreTg/Srsf2fl/fl HSPCs had reduced colony-forming capacities in vitro relative to control, suggesting severe functional defects in homozygous mutants that are otherwise not observed in heterozygous mice.
The above data identify that both Srsf1 and Srsf2 are essential for normal hematopoietic function in both embryonic and adult life. Although SRSF1 and SRSF2 have been suggested to regulate splicing by binding to similar sequences of pre-mRNA to promote splicing, here we find that both are absolutely required for hematopoiesis in a haplosufficient, non-overlapping manner. We are now performing transcriptomic analyses to delineate the common as well as distinct molecular targets of how SR proteins regulate HSPC function.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal