Abstract
Introduction
Diffuse large B cell lymphoma (DLBCL), the most common lymphoma world-wide, is strikingly heterogeneous. This heterogeneity creates a daunting challenge for conducting well-powered studies connecting molecular features to clinical outcome. Not only is the association of genetic mutations with clinical outcome in DLBCL mostly unknown, the relative importance of other well-described features, such as MYC and BCL2 translocation/expression and cell of origin based subsets (ABC and GCB DLBCL), is difficult to interpret due to conflicting reports.
We sought to comprehensively define the spectrum of genetic mutations and their association with clinical outcome in DLBCL. Our calculations indicated that 500 tumor-normal pairs would provide 95% power to define mutations occurring in at least 5% of patients, and that 800 cases would be required to define the clinical correlations with cross-validation.
Methods
We enrolled 1001 de novo DLBCL patients, with complete IPI and survival data, who were treated uniformly with standard rituximab and anthracycline containing regimens. All tumors were subjected to whole exome and transcriptome sequencing (RNAseq), as well as SNP arrays to confirm genetic alterations. ABC (38%) and GCB DLBCL (36%) subtypes were defined using microarrays and RNAseq in these patients to examine subgroup-based differences in mutations and outcome. MYC and BCL2 expression were quantified separately.
Results
Gene discovery analysis of somatic mutations and copy number alterations in exome sequencing data from 502 tumor-normal pairs of DLBCL identified 197 recurrently mutated genes, including 155 genes previously identified to be mutated in DLBCLs. In addition, our study uncovered 42 novel driver genes in DLBCL (e.g. BTK, SPEN, CD70). Exome sequencing results were validated by Sanger sequencing of 1120 variants with over 90% concordance. We also identified copy number alterations in these genes, with strong agreement (90%) of amplifications and/or deletions to those detected on Illumina high resolution SNP microarrays.
These 197 genes were found to comprise 15 functionally related subnetworks, including those related to histone modification, NFkB, B cell receptor, PI3K and cell cycle (Figure 1). Within each subnetwork, the vast majority of the gene alterations occurred in a mutually exclusive (P<10-3) fashion in patterns consistent with their described functions within the subnetworks. For instance, among genes comprising the NFkB subnetwork, positive regulators of the pathway such as MYD88 and CARD11 showed activating patterns (copy number gains and recurrent hotspot mutations), whereas negative regulators such as TNFAIP3, NFKBIE, and NFKBIA were inactivated through genetic deletions or frequent nonsense or frameshift mutations.
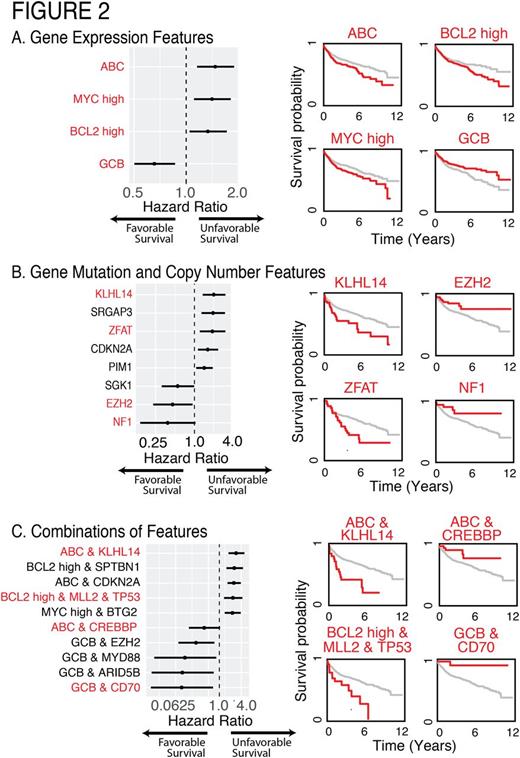
We examined the associations between the mutations and clinical outcome in all 1001 patients. All survival analyses were conducted using nearly equally split training and validation sets, corrected for multiple comparisons with significance of P<0.01 in the validation set (Figure 2). The cell of origin classification was strongly associated with survival in our cases and was independent of MYC and BCL2 co-expression, which was separately associated with survival (Figure 2A). Figure 2B shows hazard ratios for select genes, as well as associated Kaplan-Meier survival curves for a subset of those genes. We further identified combinations of different genetic and expression features that point to context dependence for survival associations (Figure 2C). For instance, mutations in KLHL14 were associated with a particularly poor prognosis in ABC DLBCL, while CREBBP mutations in ABC DLBCL patients were associated with better prognosis than average GCB DLBCLs. Mutations in EZH2 and CD70 were associated with a highly favorable prognosis within the GCB DLBCL subgroup. TP53 mutations were found to be prognostic only in the presence of MLL2 mutations and high BCL2 expression. Importantly, these risk groups are mutually exclusive and inform clinical outcome significantly better than existing metrics.
Conclusion
To our knowledge, this is the largest whole exome sequencing study in any single cancer. Our study answers many long-standing questions in the disease, informing a comprehensive understanding of genetic drivers of DLBCL, their organization into pathways, and their relationship to clinical outcome.
Leppä:Roche: Consultancy, Honoraria, Other: Travel expenses, Research Funding; Takeda Pharmaceuticals: Consultancy, Honoraria, Other: Travel expenses; Janssen: Consultancy, Research Funding; CTI Bio Pharma: Consultancy; Mundipharma: Research Funding; Bayer: Other: Travel expenses, Research Funding. Flowers:ECOG: Research Funding; Gilead: Consultancy, Research Funding; Millenium/Takeda: Research Funding; Pharmacyclics, LLC, an AbbVie Company: Research Funding; TG Therapeutics: Research Funding; Mayo Clinic: Research Funding; NIH: Research Funding; Infinity: Research Funding; AbbVie: Research Funding; Genentech: Consultancy, Research Funding; Roche: Consultancy, Research Funding; Acerta: Research Funding. Hsi:Seattle Genetics: Honoraria, Speakers Bureau; HTG Molecular Diagnostics: Consultancy, Honoraria; Eli Lilly: Research Funding; Cellerant Therapeutics: Honoraria, Research Funding; Abbvie: Honoraria, Research Funding. Evens:Takeda: Other: Advisory board. Reddy:GILEAD: Membership on an entity's Board of Directors or advisory committees; INFINITY: Membership on an entity's Board of Directors or advisory committees; celgene: Membership on an entity's Board of Directors or advisory committees; KITE: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal