To the editor:
The multiple zinc finger-containing transcription factor, IKAROS/IKZF1, has multifaceted roles in hematopoiesis.1 Numerous studies using both human primary hematopoietic cells and mouse models have suggested that IKZF1 plays a key role in erythropoiesis and the fetal-to-adult hemoglobin (Hb) switch.2-7 Even with partial suppression of IKZF1, fetal Hb (HbF) silencing appeared to be impaired in these model systems.4,7 In addition, extensive studies of various mouse models have demonstrated that Ikzf1 is necessary for hematopoietic stem cell (HSC) maintenance, lymphoid-priming in early multipotential hematopoietic progenitors, B- and T-lymphopoiesis, and suppression of specific myeloid lineages. Ikzf1 knockout mice have a 30- to 40-fold reduction in HSCs, an absence of all B- and T-lymphoid precursors, and an increase in the number of myeloid precursors, including those giving rise to megakaryocytes, basophils, and neutrophils.1,8-10 Ongoing work is focused on understanding the mechanisms through which IKZF1 is able to mediate these various activities during hematopoiesis through association with chromatin regulatory complexes.1,11 However, recent findings have also highlighted key differences in aspects of hematopoiesis observed in humans compared with various model systems.12-15 Importantly, a study in primary human hematopoietic cells has shown that suppression of IKZF1 does not alter HbF silencing and has little effect on erythropoiesis,16 in contrast with other earlier studies.4,7 Thus, analysis of human IKZF1 mutations in vivo would enhance our understanding of this gene’s function, similar to the manner in which other human genetic studies have provided key insights into hematopoiesis and Hb gene regulation.15,17-21 For example, haploinsufficiency of the multiple zinc finger-containing transcription factor BCL11A in humans is sufficient to allow high-level persistence of HbF, validating this factor as a key therapeutic target for HbF induction to treat patients with sickle cell disease (SCD) and β-thalassemia.17,18,22-24
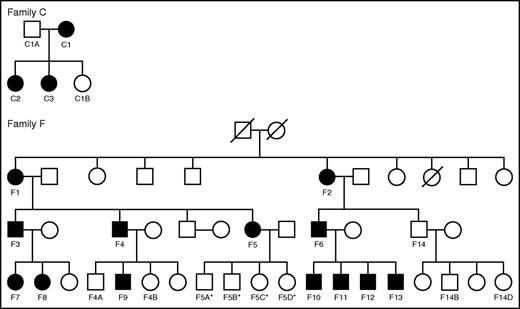
Recently, an autosomal dominant form of a common variable immunodeficiency (CVID)-like syndrome with progressive B lymphopenia and an increased risk of hematologic malignancies was associated with heterozygous loss-of-function mutations in IKZF1.25 Despite an extensive analysis of immunologic parameters in these individuals, hematologic phenotypes were not obviously apparent or described. We therefore performed hematologic analyses (including complete blood counts [CBCs], white blood cell [WBC] subset assessments, and Hb subtype quantification) on a total of 16 affected individuals carrying IKZF1 mutations and 11 unaffected family members from 2 separate pedigrees (Table 1; Figure 1). Approval for this study was obtained from the Institutional Review Boards at the National Institutes of Health, the University of Utah, and Boston Children’s Hospital. Informed consent for this study was provided according to the Declaration of Helsinki. Family C has a c.500A>G mutation resulting in the H167R loss-of-function missense mutation, whereas Family F has a 4.7 Mb heterozygous deletion on chromosome 7 involving one IKZF1 allele (Figure 1).25
Hematologic parameters for individuals from Families C and F
| . | . | . | Family C . | . | . | Family F . | Normal range . | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.500A→G (p.H167R) . | 4.7-Mb deletion on chromosome 7 . | |||||||||||||||||||||||||||
| Subject | C1 | C1A | C2 | C3 | C1B | F1 | F2 | F3 | F4 | F4A | F4B | F5 | F5A | F5B | F5C | F5D | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F14B | F14D | — |
| Sex | F | M | F | F | F | F | F | M | M | M | F | F | M | M | F | F | M | F | F | M | M | M | M | M | M | M | F | — |
| Age | 49 | 47 | 18 | 16 | 14 | 64 | 71 | 39 | 37 | 10 | 6 | 35 | 11 | 3 | 12 | 6 | 48 | 14 | 10 | 8 | 23 | 21 | 19 | 16 | 44 | 12 | 11 | — |
| Affected/ Unaffected (A/U) | A | U | A | A | U | A | A | A | A | U | U | A | U* | U* | U* | U* | A | A | A | A | A | A | A | A | U | U | U | — |
| WBC, K/μL | 9.01 | 6.34 | 7.58 | 9.68 | 13.08 | 6.10 | 3.95 | 4.70 | 6.45 | 7.04 | 8.36 | 8.62 | 8.4 | 9.84 | 10.7 | 7.32 | 5.40 | 4.81 | 5.95 | 4.74 | 7.87 | 8.9 | 8.68 | 6.98 | 10.33 | 6.32 | 5.67 | 3.98-10.04 |
| RBC, M/μL | 4.80 | 5.13 | 5.08 | 4.80 | 4.63 | 3.40 | 4.51 | 4.79 | 4.65 | 4.14 | 3.69 | 4.31 | 4.7 | 3.7 | 4.71 | 4.33 | 5.5 | 4.98 | 5.02 | 4.33 | 5.19 | 5.18 | 4.97 | 5.13 | 5.06 | 4.74 | 4.35 | 3.93-5.22 |
| Hb, g/dL | 13.9 | 16.3 | 13.1 | 14.6 | 13.7 | 10.2 | 13.9 | 15.1 | 14.8 | 12.2 | 10.5 | 13.0 | 13.6 | 10.5 | 13.7 | 12.5 | 16.1 | 14.7 | 13.9 | 12.1 | 16.9 | 17.4 | 16.0 | 16.6 | 15.2 | 14.0 | 13.2 | 11.0-17.0 |
| HCT, % | 41.5 | 46.9 | 40.7 | 42.1 | 40.3 | 32.3 | 43.1 | 45.8 | 45.0 | 37.5 | 33.2 | 42.4 | 42.7 | 33.2 | 42.8 | 39.1 | 50.6 | 46.9 | 43.8 | 38.8 | 50.1 | 52.8 | 47.9 | 49.9 | 49.1 | 44.6 | 41.1 | 34.0-50.0 |
| MCV, fL | 86.5 | 91.4 | 80.1 | 87.7 | 87.0 | 95.0 | 95.6 | 95.6 | 96.8 | 90.6 | 90.0 | 98.4 | 90.9 | 89.7 | 90.9 | 90.3 | 92.0 | 94.2 | 87.3 | 89.6 | 96.5 | 101.9 | 96.4 | 97.3 | 97.0 | 94.1 | 94.5 | 79.4-96.0 |
| MCH, pg | 29.0 | 31.8 | 25.8 | 30.4 | 29.6 | 30.0 | 30.8 | 31.5 | 31.8 | 29.5 | 28.5 | 30.2 | 28.9 | 28.4 | 29.1 | 28.9 | 29.3 | 29.5 | 27.7 | 27.9 | 32.6 | 33.6 | 32.2 | 32.4 | 30.0 | 29.5 | 30.3 | 25.6-33.0 |
| MCHC, g/dL | 33.5 | 34.8 | 32.2 | 34.7 | 34.0 | 31.6 | 32.3 | 33.0 | 32.9 | 32.5 | 31.6 | 30.7 | 31.9 | 31.6 | 32.0 | 32.0 | 31.8 | 31.3 | 31.7 | 31.2 | 33.7 | 33.0 | 33.4 | 33.3 | 31.0 | 31.4 | 32.1 | 32.2-35.5 |
| RDW, % | 12.7 | 12.6 | 14.1 | 13.1 | 12.7 | 14.1 | 14.9 | 13.8 | 14.1 | 12.9 | 13.3 | 13.4 | 13.3 | 14.4 | 13.2 | 13.4 | 14.6 | 13.5 | 14.3 | 13.0 | 13.5 | 13.3 | 14.3 | 14.0 | 12.7 | 13.4 | 12.3 | 11.7-14.4 |
| Platelet count, K/μL | 361 | 209 | 279 | 231 | 329 | 254 | 185 | 225 | 179 | 285 | 291 | 350 | 333 | 448 | 430 | 371 | 207 | 273 | 281 | 266 | 230 | 354 | 315 | 288 | 344 | 416 | 353 | 173-369 |
| MPV, fL | 10.0 | 11.0 | 10.2 | 10.0 | 8.9 | 11.4 | 10.6 | 11.0 | 11.8 | 10.0 | 10.08 | 10.4 | 10.9 | 10.2 | 10.0 | 10.1 | 9.5 | 9.8 | 10.1 | 10.5 | 9.6 | 8.6 | 9.2 | 8.8 | 11.0 | 11.0 | 11.2 | 9.4-12.3 |
| Nucleated RBCs, K/μL | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.0 | 0.00 | 0.02 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.00-0.01 |
| Neutrophils, K/μL | 4.51 | 3.66 | 3.84 | 4.25 | 10.15 | 3.43 | 1.45 | 3.13 | 2.49 | 3.35 | 3.86 | 5.08 | 4.02 | 4.23 | 4.69 | 2.76 | 3.03 | 2.44 | 2.48 | 1.51 | 3.97 | 4.18 | 3.38 | 3.70 | 6.33 | 3.42 | 2.46 | 1.56-6.13 |
| Absolute lymphocytes, K/μL | 3.36 | 1.98 | 2.81 | 4.60 | 1.85 | 1.86 | 2.11 | 1.02 | 3.26 | 2.76 | 3.78 | 2.67 | 3.29 | 4.82 | 4.81 | 3.66 | 1.73 | 1.75 | 2.51 | 2.65 | 2.87 | 3.86 | 4.10 | 2.21 | 2.78 | 2.00 | 2.52 | 1.18-3.74 |
| Absolute monocytes, K/μL | 0.77 | 0.49 | 0.74 | 0.78 | 0.88 | 0.64 | 0.37 | 0.35 | 0.58 | 0.47 | 0.52 | 0.59 | 0.62 | 0.65 | 0.91 | 0.63 | 0.52 | 0.44 | 0.56 | 0.46 | 0.62 | 0.6 | 0.81 | 0.74 | 0.69 | 0.52 | 0.49 | 0.24-0.86 |
| Absolute eosinophils, K/μL | 0.21 | 0.13 | 0.09 | 0.00 | 0.08 | 0.12 | <0.03 | 0.14 | 0.05 | 0.42 | 0.15 | 0.19 | 0.39 | 0.07 | 0.17 | 0.19 | <0.03 | 0.12 | 0.35 | 0.06 | 0.26 | 0.17 | 0.25 | 0.20 | 0.41 | 0.31 | 0.17 | 0.04-0.36 |
| Absolute basophils, K/μL | 0.09 | 0.05 | 0.05 | 0.05 | 0.06 | 0.03 | <0.03 | 0.04 | 0.05 | 0.04 | 0.03 | 0.06 | 0.05 | 0.04 | 0.07 | 0.06 | 0.09 | 0.04 | 0.04 | 0.04 | 0.12 | 0.06 | 0.12 | 0.12 | 0.08 | 0.06 | <0.03 | 0.00-0.10 |
| Reticulocyte, % | — | — | — | — | — | 1.4 | 1.3 | 1.0 | 1.1 | 0.9 | 0.9 | 1.5 | 1.2 | 1.7 | 1.4 | 1.4 | 1.8 | 0.9 | 0.8 | 0.8 | 2.5 | 3.3 | 2.7 | 2.2 | 1.5 | 1.4 | 1.3 | 0.5-2.0 |
| Absolute reticulocytes, K/μL | — | — | — | — | — | 46.9 | 60.0 | 46.5 | 49.3 | 37.7 | 33.6 | 64.7 | 56.6 | 62.9 | 64.1 | 60.6 | 96.8 | 44.8 | 41.2 | 34.6 | 126.0 | 171.5 | 134.2 | 113.9 | 77.4 | 68.3 | 56.1 | — |
| HbF, % | 1.5 | 1.1 | 1.1 | 0.5 | 0.8 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | 0.2 | 1.5 | 0.8 | 0.3 | 0.2 | 0.2 | 0.4 | 0.2 | 0.2 | 0.7 | 0.2 | 0.2 | 0.3 | 0.3 | 0.9 | <2.0 |
| HbA2, % | 2.2 | 2.5 | 2.1 | 2.1 | 2.7 | 2.8 | 2.9 | 2.9 | 3.1 | 2.8 | 3.2 | 2.7 | 3.2 | 3.1 | 3.0 | 3.0 | 3.0 | 2.8 | 2.7 | 3.2 | 3.0 | 3.1 | 2.9 | 2.9 | 3.0 | 2.9 | 3.1 | <3.5 |
| . | . | . | Family C . | . | . | Family F . | Normal range . | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.500A→G (p.H167R) . | 4.7-Mb deletion on chromosome 7 . | |||||||||||||||||||||||||||
| Subject | C1 | C1A | C2 | C3 | C1B | F1 | F2 | F3 | F4 | F4A | F4B | F5 | F5A | F5B | F5C | F5D | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F14B | F14D | — |
| Sex | F | M | F | F | F | F | F | M | M | M | F | F | M | M | F | F | M | F | F | M | M | M | M | M | M | M | F | — |
| Age | 49 | 47 | 18 | 16 | 14 | 64 | 71 | 39 | 37 | 10 | 6 | 35 | 11 | 3 | 12 | 6 | 48 | 14 | 10 | 8 | 23 | 21 | 19 | 16 | 44 | 12 | 11 | — |
| Affected/ Unaffected (A/U) | A | U | A | A | U | A | A | A | A | U | U | A | U* | U* | U* | U* | A | A | A | A | A | A | A | A | U | U | U | — |
| WBC, K/μL | 9.01 | 6.34 | 7.58 | 9.68 | 13.08 | 6.10 | 3.95 | 4.70 | 6.45 | 7.04 | 8.36 | 8.62 | 8.4 | 9.84 | 10.7 | 7.32 | 5.40 | 4.81 | 5.95 | 4.74 | 7.87 | 8.9 | 8.68 | 6.98 | 10.33 | 6.32 | 5.67 | 3.98-10.04 |
| RBC, M/μL | 4.80 | 5.13 | 5.08 | 4.80 | 4.63 | 3.40 | 4.51 | 4.79 | 4.65 | 4.14 | 3.69 | 4.31 | 4.7 | 3.7 | 4.71 | 4.33 | 5.5 | 4.98 | 5.02 | 4.33 | 5.19 | 5.18 | 4.97 | 5.13 | 5.06 | 4.74 | 4.35 | 3.93-5.22 |
| Hb, g/dL | 13.9 | 16.3 | 13.1 | 14.6 | 13.7 | 10.2 | 13.9 | 15.1 | 14.8 | 12.2 | 10.5 | 13.0 | 13.6 | 10.5 | 13.7 | 12.5 | 16.1 | 14.7 | 13.9 | 12.1 | 16.9 | 17.4 | 16.0 | 16.6 | 15.2 | 14.0 | 13.2 | 11.0-17.0 |
| HCT, % | 41.5 | 46.9 | 40.7 | 42.1 | 40.3 | 32.3 | 43.1 | 45.8 | 45.0 | 37.5 | 33.2 | 42.4 | 42.7 | 33.2 | 42.8 | 39.1 | 50.6 | 46.9 | 43.8 | 38.8 | 50.1 | 52.8 | 47.9 | 49.9 | 49.1 | 44.6 | 41.1 | 34.0-50.0 |
| MCV, fL | 86.5 | 91.4 | 80.1 | 87.7 | 87.0 | 95.0 | 95.6 | 95.6 | 96.8 | 90.6 | 90.0 | 98.4 | 90.9 | 89.7 | 90.9 | 90.3 | 92.0 | 94.2 | 87.3 | 89.6 | 96.5 | 101.9 | 96.4 | 97.3 | 97.0 | 94.1 | 94.5 | 79.4-96.0 |
| MCH, pg | 29.0 | 31.8 | 25.8 | 30.4 | 29.6 | 30.0 | 30.8 | 31.5 | 31.8 | 29.5 | 28.5 | 30.2 | 28.9 | 28.4 | 29.1 | 28.9 | 29.3 | 29.5 | 27.7 | 27.9 | 32.6 | 33.6 | 32.2 | 32.4 | 30.0 | 29.5 | 30.3 | 25.6-33.0 |
| MCHC, g/dL | 33.5 | 34.8 | 32.2 | 34.7 | 34.0 | 31.6 | 32.3 | 33.0 | 32.9 | 32.5 | 31.6 | 30.7 | 31.9 | 31.6 | 32.0 | 32.0 | 31.8 | 31.3 | 31.7 | 31.2 | 33.7 | 33.0 | 33.4 | 33.3 | 31.0 | 31.4 | 32.1 | 32.2-35.5 |
| RDW, % | 12.7 | 12.6 | 14.1 | 13.1 | 12.7 | 14.1 | 14.9 | 13.8 | 14.1 | 12.9 | 13.3 | 13.4 | 13.3 | 14.4 | 13.2 | 13.4 | 14.6 | 13.5 | 14.3 | 13.0 | 13.5 | 13.3 | 14.3 | 14.0 | 12.7 | 13.4 | 12.3 | 11.7-14.4 |
| Platelet count, K/μL | 361 | 209 | 279 | 231 | 329 | 254 | 185 | 225 | 179 | 285 | 291 | 350 | 333 | 448 | 430 | 371 | 207 | 273 | 281 | 266 | 230 | 354 | 315 | 288 | 344 | 416 | 353 | 173-369 |
| MPV, fL | 10.0 | 11.0 | 10.2 | 10.0 | 8.9 | 11.4 | 10.6 | 11.0 | 11.8 | 10.0 | 10.08 | 10.4 | 10.9 | 10.2 | 10.0 | 10.1 | 9.5 | 9.8 | 10.1 | 10.5 | 9.6 | 8.6 | 9.2 | 8.8 | 11.0 | 11.0 | 11.2 | 9.4-12.3 |
| Nucleated RBCs, K/μL | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.0 | 0.00 | 0.02 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.00-0.01 |
| Neutrophils, K/μL | 4.51 | 3.66 | 3.84 | 4.25 | 10.15 | 3.43 | 1.45 | 3.13 | 2.49 | 3.35 | 3.86 | 5.08 | 4.02 | 4.23 | 4.69 | 2.76 | 3.03 | 2.44 | 2.48 | 1.51 | 3.97 | 4.18 | 3.38 | 3.70 | 6.33 | 3.42 | 2.46 | 1.56-6.13 |
| Absolute lymphocytes, K/μL | 3.36 | 1.98 | 2.81 | 4.60 | 1.85 | 1.86 | 2.11 | 1.02 | 3.26 | 2.76 | 3.78 | 2.67 | 3.29 | 4.82 | 4.81 | 3.66 | 1.73 | 1.75 | 2.51 | 2.65 | 2.87 | 3.86 | 4.10 | 2.21 | 2.78 | 2.00 | 2.52 | 1.18-3.74 |
| Absolute monocytes, K/μL | 0.77 | 0.49 | 0.74 | 0.78 | 0.88 | 0.64 | 0.37 | 0.35 | 0.58 | 0.47 | 0.52 | 0.59 | 0.62 | 0.65 | 0.91 | 0.63 | 0.52 | 0.44 | 0.56 | 0.46 | 0.62 | 0.6 | 0.81 | 0.74 | 0.69 | 0.52 | 0.49 | 0.24-0.86 |
| Absolute eosinophils, K/μL | 0.21 | 0.13 | 0.09 | 0.00 | 0.08 | 0.12 | <0.03 | 0.14 | 0.05 | 0.42 | 0.15 | 0.19 | 0.39 | 0.07 | 0.17 | 0.19 | <0.03 | 0.12 | 0.35 | 0.06 | 0.26 | 0.17 | 0.25 | 0.20 | 0.41 | 0.31 | 0.17 | 0.04-0.36 |
| Absolute basophils, K/μL | 0.09 | 0.05 | 0.05 | 0.05 | 0.06 | 0.03 | <0.03 | 0.04 | 0.05 | 0.04 | 0.03 | 0.06 | 0.05 | 0.04 | 0.07 | 0.06 | 0.09 | 0.04 | 0.04 | 0.04 | 0.12 | 0.06 | 0.12 | 0.12 | 0.08 | 0.06 | <0.03 | 0.00-0.10 |
| Reticulocyte, % | — | — | — | — | — | 1.4 | 1.3 | 1.0 | 1.1 | 0.9 | 0.9 | 1.5 | 1.2 | 1.7 | 1.4 | 1.4 | 1.8 | 0.9 | 0.8 | 0.8 | 2.5 | 3.3 | 2.7 | 2.2 | 1.5 | 1.4 | 1.3 | 0.5-2.0 |
| Absolute reticulocytes, K/μL | — | — | — | — | — | 46.9 | 60.0 | 46.5 | 49.3 | 37.7 | 33.6 | 64.7 | 56.6 | 62.9 | 64.1 | 60.6 | 96.8 | 44.8 | 41.2 | 34.6 | 126.0 | 171.5 | 134.2 | 113.9 | 77.4 | 68.3 | 56.1 | — |
| HbF, % | 1.5 | 1.1 | 1.1 | 0.5 | 0.8 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | 0.2 | 1.5 | 0.8 | 0.3 | 0.2 | 0.2 | 0.4 | 0.2 | 0.2 | 0.7 | 0.2 | 0.2 | 0.3 | 0.3 | 0.9 | <2.0 |
| HbA2, % | 2.2 | 2.5 | 2.1 | 2.1 | 2.7 | 2.8 | 2.9 | 2.9 | 3.1 | 2.8 | 3.2 | 2.7 | 3.2 | 3.1 | 3.0 | 3.0 | 3.0 | 2.8 | 2.7 | 3.2 | 3.0 | 3.1 | 2.9 | 2.9 | 3.0 | 2.9 | 3.1 | <3.5 |
HCT, hematocrit; MCH, mean corpuscular Hb; MCHC, MHC concentration; MCV, mean corpuscular volume; MPV, mean platelet volume; RBC, red blood cell; RDW, RBC distribution width.
Pedigrees of families with IKZF1 mutations. Families C and F are shown. Female (circle) and male (square) individuals are shown in the 2 pedigrees and the shaded shapes indicate individuals who are affected by a CVID-like syndrome due to heterozygous IKZF1 mutations. Individuals with hematologic phenotypes available are labeled in the pedigrees and correspond to the labeling shown in Table 1. *Indicates individuals who are clinically unaffected, but for whom molecular testing results are not yet available.
Pedigrees of families with IKZF1 mutations. Families C and F are shown. Female (circle) and male (square) individuals are shown in the 2 pedigrees and the shaded shapes indicate individuals who are affected by a CVID-like syndrome due to heterozygous IKZF1 mutations. Individuals with hematologic phenotypes available are labeled in the pedigrees and correspond to the labeling shown in Table 1. *Indicates individuals who are clinically unaffected, but for whom molecular testing results are not yet available.
CBC results revealed that nearly all blood cell parameters were within the normal range in both affected individuals and unaffected family members (Table 1). No defects in either RBC, platelet, or WBC subtype numbers or parameters were observed with the exception of very mild macrocytosis and reticulocytosis observed in siblings F10-F13. Given the absence of similar findings in other family members carrying the identical IKZF1 deletion, we suspect that these altered parameters may be due to a different genetic or environmental etiology (Table 1). Although we cannot entirely rule out whether subtle defects in precursors may be present in the bone marrow of all of the affected individuals in Families C and F, blood counts are remarkably sensitive indicators of variation in hematopoiesis. We do note a trend toward higher basophil counts in several affected individuals (eg, C1, F6, F10, F12, and F13), consistent with the known role of IKZF1 in restricting basophil production.9 Other myeloid lineages appeared to be within the normal ranges consistently in all individuals. Importantly, we observed no significant alteration in Hb subtype levels, and specifically in HbF levels, in any of the affected individuals (Table 1). Although greater suppression of IKZF1 may be necessary to observe altered hematopoiesis or globin gene regulation, our results suggest that heterozygous loss-of-function, resulting in a reduced level of IKZF1 sufficient to alter B-lymphocyte production and function, is insufficient to alter other hematologic parameters or HbF silencing. This phenotype contrasts with those observed for mutations in other related factors such as BCL11A, to which IKZF1 has been suggested to bind in erythroid cells.7 Haploinsufficiency for BCL11A can result in substantial persistence of HbF of >10%.17,18,22
It is noteworthy that we were able to evaluate 2 families with different types of heterozygous IKZF1 mutations: a missense change in Family C and a complete gene deletion in Family F. Because IKZF1 predominantly acts as a homodimeric transcription factor, affected members in Family F are expected to produce only ∼50% of IKZF1 wild-type (WT)/WT homodimers, whereas affected individuals in Family C should make ∼25% of such dimers (plus ∼25% of mutant/mutant and ∼50% of WT/mutant dimers).25 Thus, the normal hematologic parameters and similar levels of Hb subtypes observed in the 2 families suggest that although ∼50% of WT/WT IKZF1 dimers are not sufficient to support physiologic B-cell development and function, ∼25% of them are enough to ensure otherwise normal hematopoiesis.
More generally, our analysis emphasizes the utility of in-depth human genetic studies and phenotyping to validate the role of key regulators in human hematopoiesis and Hb gene regulation. Indeed, given the differences in hematopoiesis between humans and the various models systems employed, it is likely that many other examples of variation in phenotypes and the necessity of particular genes may be observed.13,14 For example, although heterozygous Bcl11a mutations in mice cause immunologic defects, this was not observed in humans with BCL11A haploinsufficiency.17,22 With considerable efforts being dedicated to induce HbF for therapeutic purposes to treat SCD and β-thalassemia,24 there is a need to not only rely on animal and cell culture models, which have tremendous value and provide considerable insight, but to attempt to validate findings in vivo in humans whenever possible. Emerging biobanks and precision medicine efforts are likely to provide incredibly valuable resources to help identify relevant and effective therapeutic opportunities. We emphasize the importance of careful and thorough observations of human subjects to validate and gain insight into hematopoiesis, based on findings from valuable model systems.
Authorship
Acknowledgments: The authors are grateful to the families described here for their willingness to participate in this study, and the members of the Sankaran laboratory for valuable comments and suggestions.
This work was supported by grants from the National Institutes of Health (National Heart, Lung, and Blood Institute U01 HL117720 and R33 HL120791; and National Institute of Diabetes and Digestive and Kidney Diseases R01 DK103794) (V.G.S.), as well as the National Institute of Allergy and Infectious Diseases (R03 AI113631) (A.K., K.V.V., and H.R.H.).
Contribution: N.A., C.F., H.R.H., S.D.R., and V.G.S. designed the research and wrote the manuscript with input from A.K., A.A.S., J.H.-R., and K.V.V.; and all authors gathered and analyzed clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vijay G. Sankaran, Division of Hematology/Oncology, Boston Children’s Hospital, 3 Blackfan Cir, CLS 03001, Boston, MA 02115; e-mail: sankaran@broadinstitute.org.
References
Author notes
N.A. and C.F. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal