Abstract
Hairy cell leukemia (HCL) is a distinct clinicopathological entity whose underlying genetic lesion has remained a mystery for over half a century. The BRAF V600E mutation is now recognized as the causal genetic event of HCL because it is somatic, present in the entire tumor clone, detectable in almost all cases at diagnosis (encompassing the whole disease spectrum), and stable at relapse. BRAF V600E leads to the constitutive activation of the RAF-MEK-extracellular signal-regulated kinase (ERK) signaling pathway which represents the key event in the molecular pathogenesis of HCL. KLF2 and CDNK1B (p27) mutations may cooperate with BRAF V600E in promoting leukemic transformation. Sensitive molecular assays for detecting BRAF V600E allow HCL (highly responsive to purine analogs) to be better distinguished from HCL-like disorders, which are treated differently. In vitro preclinical studies on purified HCL cells proved that BRAF and MEK inhibitors can induce marked dephosphorylation of MEK/ERK, silencing of RAF-MEK-ERK pathway transcriptional output, loss of the HCL-specific gene expression profile signature, change of morphology from “hairy” to “smooth,” and eventually apoptosis. The overall response rate of refractory/relapsed HCL patients to the BRAF inhibitor vemurafenib approached 100%, with 35% to 40% complete remissions (CRs). The median relapse free-survival was about 19 months in patients who had achieved CR and 6 months in those who had obtained a partial response. Future therapeutic perspectives include: (1) combining BRAF inhibitors with MEK inhibitors or immunotherapy (anti-CD20 monoclonal antibody) to increase the percentage of CRs and (2) better understanding of the molecular mechanisms underlying resistance of HCL cells to BRAF inhibitors.
Introduction
Hairy cell leukemia (HCL) is a rare mature B-cell lymphoid malignancy that is listed as a distinct entity in the World Health Organization (WHO) classification of lymphohemopoietic tissues.1 Characteristically, HCL occurs in older patients (median age about 60 years) with male predominance (male:female ratio = 4:1) and usually presents with pancytopenia and monocytopenia associated with hepatosplenomegaly in the absence of lymph node enlargement.1 This clinical presentation reflects the marked infiltration of bone marrow (BM), spleen, and liver (usually with sparing of lymph nodes) by leukemic cells with wide cytoplasm and long, slender cell-surface projections. This hairy morphology can be appreciated only by careful examination of smear preparations and gives the disease its vivid, descriptive name.2 The HCL cells are positive for the B-cell–associated antigens CD19, CD20, and CD22 and typically coexpress the CD103, CD25, and CD11c molecules.1 As compared to other lymphoid malignancies, HCL responds well to therapy with purine analogs (ie, cladribine and pentostatin).3
In spite of such peculiar clinicopathological features, the genetic lesion underlying HCL has remained a mystery for over half a century since the original description of the disease in 1958 under the term of leukemic reticuloendotheliosis.4 A major obstacle to investigating the genome and functional features of HCL has been the inability to collect enough numbers of leukemic cells for analysis. In fact, HCL patients usually present with pancytopenia and a low number of circulating tumor cells; moreover, the BM aspiration frequently results in dry-tap and leukemic cells cannot usually be collected from markedly enlarged spleen because splenectomy is rarely performed in the purine analogs era. Additional problems in the genomic and functional studies of HCL are the very low proliferative index of leukemic cells and the lack of true HCL cell lines5,6 as well as of faithful animal models of the disease.7
Cytogenetic/fluorescence in situ hybridization, comparative genomic hybridization, and microarray single-nucleotide polymorphism genotyping did not identify chromosomal translocations or copy-number aberrations consistently associated with HCL at significant frequencies (≥10% of cases).8-12 Both gene expression (GEP)13 and microRNA14 profiles revealed a unique molecular signature that, at least in part, may explain some distinctive functional features of HCL cells, such as the hairy morphology, the associated BM fibrosis, the typical adhesion properties, and the selective homing to particular anatomical sites.15 High-density genome-wide single-nucleotide polymorphism genotyping clearly showed that HCL has a remarkable stable genome,16 as compared with other B-cell neoplasms. However, none of the above approaches was able to unravel any recurrent genetic alteration responsible for the disease.
The BRAF V600E mutation is the causal genetic event in HCL
Using a whole-exome sequencing approach, in 2011 we discovered the BRAF V600E mutation as the causal genetic event of HCL.17 The characteristics of this mutation and its role in the pathogenesis of HCL are explained in more detail in the following sections.
Unique characteristics of the BRAF V600E mutation in HCL
BRAF V600E is a point mutation (substitution of a thymine with adenine at position 1799 on exon 15) that results in the change of amino acid 600 from valine (V) to glutamate (E). The BRAF V600E mutation causes constitutive activation of the RAS-RAF-MEK-ERK signaling pathway, independent of extracellular signals (see next section). Notably, among B-cell neoplasms, HCL is the only pathological condition in which the molecular pathogenesis is mediated by an activating point mutation of a kinase-encoding gene.17 BRAF mutations have been detected in >97% of HCL patients so far investigated.17-36
Only 1 study26 showed a lower frequency of BRAF mutations (79%) in 53 bona fide HCL patients studied. However, 5 of 11 BRAF− cases had a IGHV4-34 rearrangement that is in general absent in classical HCL. Unlike that observed in other BRAF-mutated tumors that may carry mutations other than V600E (eg, V600K and V600R in melanoma), virtually all HCL patients were V600E+.17-36 In 2 V600E− HCL patients, novel, potentially functionally relevant mutations in exon 11 (F468C and D449E) were detected.35
There are a number of findings suggesting that BRAF V600E is the disease-defining genetic event in HCL. First, this mutation, with very rare exceptions, is detectable at diagnosis in virtually all HCL patients,17-36 encompassing the whole clinical spectrum of the disease, including cases presenting with leukocytosis or without splenomegaly. Interestingly, the BRAF V600E mutation could be detected in anatomical sites unusually involved with HCL, such as lymph nodes,37 again pointing to its key role in the pathogenesis of the disease. Second, the BRAF V600E mutation is very stable over time, being consistently detectable at relapse, even after decades from the initial diagnosis. Third, in each individual patient, the mutation appears to be present in the entire tumor cell clone as demonstrated by the percentage of allelic variants at next-generation sequencing and by immunostaining of BM biopsies with an anti-BRAF V600E monoclonal antibody.33 Finally, among B-cell chronic lymphoproliferative disorders, the BRAF V600E mutation is very specific for HCL.17,21,38
BRAF V600E–mediated constitutive activation of the RAS-RAF-MEK-ERK pathway is the key event in the molecular pathogenesis of HCL
The BRAF protein (encoded by the BRAF proto-oncogene at chromosome 7q24) is a member of the serine–threonine kinase RAF family (comprising RAF-1/CRAF, ARAF, BRAF). BRAF is a key component of the RAS-RAF-MEK-ERK signaling pathway (Figure 1). RAF proteins share many structural and biological properties but they differ in basal kinase activity that is, in turn, depending on the phosphorylation status of these molecules. In particular, BRAF appears to have a basal kinase activity higher than CRAF because serine 445 is constitutively phosphorylated in BRAF whereas the homologous site (serine 338) in CRAF is not and needs to be phosphorylated in order to exploit maximal RAS-induced activation.39,40 This explains why a single mutation at codon 600 (V600E) results in constitutive activation of BRAF but not of CRAF41 and also why BRAF is the most frequently mutated RAF protein in human cancers.42
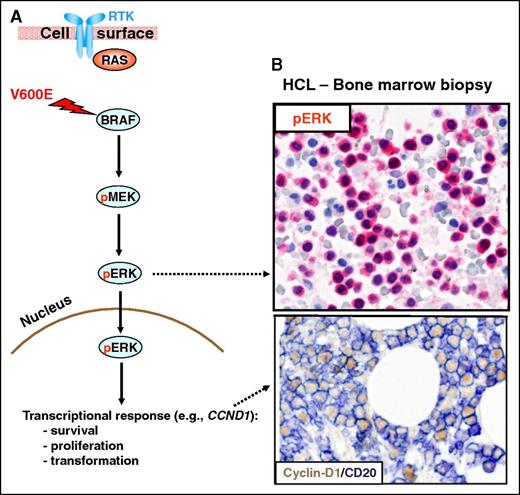
The RAS-RAF-MEK-ERK signaling pathway. (A) The RAS-RAF-MEK-ERK signaling pathway is physiologically triggered by the binding of a surface receptor tyrosine kinase (RTK) to its ligand. This activates RAS and, in turn, RAFs (BRAF and, not shown, CRAF). BRAF-CRAF heterodimers phosphorylate the MEK1 and MEK2 kinases (pMEK), which in turn phosphorylate ERK1 and ERK2 (pERK). Then, active ERKs phosphorylate several substrates in the cytoplasm (not shown) as well as in the nucleus, where they initiate a transcriptional response (eg, through the activator protein 1 transcription complex) that includes cyclin-D1 upregulation and that promotes cell survival and proliferation, as well as feedback inhibitory mechanisms (not shown) to counterregulate pathway activity. The latter, if uncontrolled, can result in neoplastic transformation. Indeed, the BRAF V600E mutation renders BRAF constitutively active independent from upstream regulatory signals and from heterodimerization with CRAF. (B) In vivo activation of the BRAF-MEK-ERK pathway in HCL patients is illustrated by the expression of pERK (red) and cyclin-D1 (nuclear, brown) by BM leukemic hairy cells (counterstained with hematoxylin in the top panel and double stained for the surface B-cell marker CD20 (surface, blue) in the bottom panel). Pictures of immunohistochemical stainings were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus).
The RAS-RAF-MEK-ERK signaling pathway. (A) The RAS-RAF-MEK-ERK signaling pathway is physiologically triggered by the binding of a surface receptor tyrosine kinase (RTK) to its ligand. This activates RAS and, in turn, RAFs (BRAF and, not shown, CRAF). BRAF-CRAF heterodimers phosphorylate the MEK1 and MEK2 kinases (pMEK), which in turn phosphorylate ERK1 and ERK2 (pERK). Then, active ERKs phosphorylate several substrates in the cytoplasm (not shown) as well as in the nucleus, where they initiate a transcriptional response (eg, through the activator protein 1 transcription complex) that includes cyclin-D1 upregulation and that promotes cell survival and proliferation, as well as feedback inhibitory mechanisms (not shown) to counterregulate pathway activity. The latter, if uncontrolled, can result in neoplastic transformation. Indeed, the BRAF V600E mutation renders BRAF constitutively active independent from upstream regulatory signals and from heterodimerization with CRAF. (B) In vivo activation of the BRAF-MEK-ERK pathway in HCL patients is illustrated by the expression of pERK (red) and cyclin-D1 (nuclear, brown) by BM leukemic hairy cells (counterstained with hematoxylin in the top panel and double stained for the surface B-cell marker CD20 (surface, blue) in the bottom panel). Pictures of immunohistochemical stainings were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus).
Under physiological conditions, the RAS-RAF-MEK-ERK signaling pathway can be activated through binding of a variety of receptors (including tyrosine kinases) with their cognate ligands43,44 (Figure 1). These interactions trigger, as first events, the recruitment of the adaptor molecules Grb2 to the inner part of the cell membrane and its binding to the guanine nucleotide-exchange factor SOS. Activated SOS, in turn, displaces guanosine diphosphate from RAS, promoting the formation of guanosine triphosphate–RAS.44 Then, RAS in its activated guanosine triphosphate–bound state recruits BRAF to the cell membrane where it is phosphorylated at threonine 599 and serine 602.44 Activated (phosphorylated) BRAF can form BRAF-CRAF heterodimers45,46 that in turn lead to phosphorylation and activation of MEK1 and MEK2.44 Finally, activated MEK1/2 phosphorylate extracellular signal-regulated kinases 1 and 2 (ERK1 and ERK2). Then, activated (phosphorylated) ERK can translocate from the cytoplasm into the nucleus where it phosphorylates the transcription factors activator protein 1 and nuclear factor of activated T cells with impact on transcription of many target genes that promote cell-cycle progression and survival.47,48
The V600E phosphomimetic substitution occurs within the BRAF-activation segment and functionally results in a marked enhancement of its kinase activity in a constitutive manner.49 BRAF mutants (especially those carrying V600E) can phosphorylate ERK as monomers and in a RAS-independent manner.49 Thus, similarly to what occurs in other BRAF-mutated tumors (including melanomas and papillary thyroid carcinomas), BRAF V600E in HCL constitutively activate the RAF-MEK-ERK pathway,17,50 leading to enhanced survival. The implication of the RAF-MEK-ERK pathway activation in the molecular pathogenesis of HCL is also supported by immunohistochemical and western blot studies showing that, in the presence of the BRAF V600E mutation, the downstream targets MEK and ERK are phosphorylated and that such a status can be reverted by specific BRAF inhibitors.17,50-52 Notably, constitutive ERK activation in HCL was previously hypothesized to provide a survival signal for the leukemic hairy cells53 in antagonism with the proapoptotic effect of p38-JNK activation (triggered by the interaction of HCL cells with the vitronectin-positive cells in the splenic red pulp).53,54 Moreover, the constitutive activation of the RAF-MEK-ERK pathway likely contributes to some of the immunophenotypic features of HCL, such as expression of cyclin D155-57 and negativity for p27.58-60
Other cooperating mutations in HCL
There is still scarce information on the genetic events that may contribute to development of HCL by cooperating with BRAF V600E. Recently, missense mutations of KLF2, a transcription factor important for the homeostasis and differentiation of peripheral B-cell subsets including marginal zone B cells, were reported in 4 of 24 HCL cases (16%), as well as in 19 of 96 splenic marginal zone lymphomas (20%), and 17 of 154 diffuse large B-cell lymphomas (11%).61 Clonal KLF2 mutants, which were mostly monoallelic, also included disruptive variants (eg, nonsense or frame-shifting alleles), and the loss-of-function nature of 1 missense mutation was experimentally documented. Soon after, using whole-exome sequencing, Dietrich et al identified recurrent mutations involving the EZH2, ARID1A, and the cell-cycle inhibitor CDKN1B (p27) genes in the genome of purine analog-refractory HCL patients.62 In particular, clonal deleterious CDKN1B mutations were detected in 13 of 81 cases (16%), strongly suggesting that they may represent driver events in the pathogenesis of HCL. Because CDNK1B is involved in the control of the cell-cycle progression, these findings suggest that alterations in the regulation of cell cycle and senescence may play an important role in the pathogenesis of HCL.
Biological implications of genomic studies
Cell of origin of HCL
GEP studies had previously shown that HCL derives from the transformation of a memory B cell.13 Similarly to normal memory B cells, HCL cells showed a conservation in proliferation, apoptosis, and DNA metabolism programs, but clearly differed from them in the expression of genes controlling cell adhesion and response to chemokines.13
More recently, it has been reported that hemopoietic stem cells (HSCs) or B-cell lymphoid progenitors from HCL patients carry the BRAF V600E mutation.7 Transplantation of BRAF V600E-mutant HSCs from an HCL patient into immunodeficient mice resulted in their stable engraftment, revealing the functional self-renewal capacity of HCL HSCs. However, none of the transplanted animals developed a full HCL phenotype.7 At present, it remains unclear whether the development of the HCL phenotype requires a permissive epigenetic background (present only in a particular stage of cell differentiation) and/or the acquirement of further genetic lesions, for example, mutations of KLF261 or CDKN1B/p27.62
Hairy morphology is dependent upon BRAF V600E-mutant–mediated activation of the RAS-RAF-MEK-ERK pathway
Recently, in vitro studies on purified HCL cells using BRAF and MEK inhibitors52,63 have clearly shown that “hairyness” represents an intrinsic property of HCL cells which is closely related to the deregulated BRAF activity, rather than reflecting the derivation from an unidentified subset of normal memory B cells with “hairy” appearance or being the result of microenvironmental stimuli (as also supported by the observation that HCL cells retain their “hairy” morphology even after several days of culture in vitro). Acquiring this unique feature may have a finalistic effect, that is, to increase the cell surface area of HCL cells in order to favor their interaction with other cells or with ligands in the extracellular matrix.
Notably, in vitro exposure of primary HCL cells to BRAF and MEK inhibitors resulted in conversion of their morphology from “hairy” to “smooth”52 (Figure 2). The inhibitors exerted their activity specifically on HCL because the morphology of primary leukemic cells from patients with HCL-like disorders was not changed.52 Analysis of how BRAF and MEK inhibitors affected the unique GEP of HCL clearly highlighted β-actin and LST1 (which were both significantly downregulated) as the most likely candidate molecules in determining the hairy morphology of HCL cells. This is consistent with a previous observation that β-actin, in its polymerized form (F-actin), is predominantly expressed in the cortical cytoskeleton of HCL, suggesting a role in sustaining the filamentous membrane projections.64 Moreover, LST1 is important in forming actin-containing long-membrane protrusions.65,66
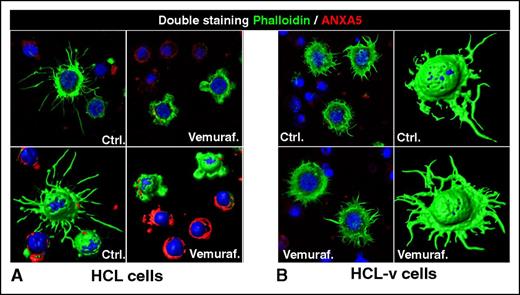
Loss of the hairy morphology upon BRAF inhibition. Leukemic cells purified from patients with (A) HCL or (B) HCL-v were exposed in vitro to vemurafenib (vemuraf.) 1 µM or drug vehicle as control (Ctrl.) for 2 or 3 days, and then costained with phalloidin (labeling in green the cytoskeleton of hairy projections rich in F-actin), Annexin-V (ANXA5, labeling in red the dying cells), and Draq5 (labeling the nucleus), followed by confocal fluorescence microscopy showing 2-dimensional images of representative cells and their corresponding 3-dimensional reconstruction. After blocking BRAF V600E with vemurafenib, HCL (but not HCL-v) cells become smoother and smaller while being still alive (ANXA5− HCL cells showing severely shortened green surface projections). This hair loss is then followed by apoptosis of HCL (but not HCL-v) cells, which become ANXA5+ (red) and lose any phalloidin-positive (green) cytoskeletal structures. These images were taken with a LSM510 laser-scanning confocal microscope (Zeiss) equipped with laser emission lines at 458, 488, 543, and 633 nm, using an objective Plan-Apocromat 63×/1.4 NA with oil immersion and LSM510 Zeiss software.
Loss of the hairy morphology upon BRAF inhibition. Leukemic cells purified from patients with (A) HCL or (B) HCL-v were exposed in vitro to vemurafenib (vemuraf.) 1 µM or drug vehicle as control (Ctrl.) for 2 or 3 days, and then costained with phalloidin (labeling in green the cytoskeleton of hairy projections rich in F-actin), Annexin-V (ANXA5, labeling in red the dying cells), and Draq5 (labeling the nucleus), followed by confocal fluorescence microscopy showing 2-dimensional images of representative cells and their corresponding 3-dimensional reconstruction. After blocking BRAF V600E with vemurafenib, HCL (but not HCL-v) cells become smoother and smaller while being still alive (ANXA5− HCL cells showing severely shortened green surface projections). This hair loss is then followed by apoptosis of HCL (but not HCL-v) cells, which become ANXA5+ (red) and lose any phalloidin-positive (green) cytoskeletal structures. These images were taken with a LSM510 laser-scanning confocal microscope (Zeiss) equipped with laser emission lines at 458, 488, 543, and 633 nm, using an objective Plan-Apocromat 63×/1.4 NA with oil immersion and LSM510 Zeiss software.
On the contrary, expression of genes that were previously hypothesized to play a role in controlling HCL morphology,15 including growth-arrest specific 7 (GAS7, required for assembling actin filaments and neurite outgrowth in neuronal cells67 ) or the CD9 molecule (which colocalizes with F-actin and is also involved in neurite growth68 ), were not affected by the exposure of HCL cells to the BRAF and MEK inhibitors.52 Thus, the role of these and other molecules previously associated with the hairy phenotype, such as pp52(LSP1)69,70 or the member of the Rho family of small GTPases (CDC42, RAC1, RHOA),71,72 in determining the morphology of HCL cells remains unclear.
Diagnostic assays inspired by genomic studies
The diagnosis of HCL is mainly based upon clinical/laboratory findings (usually splenomegaly associated with pancytopenia and monocytopenia) and the typical morphology and immunophenotype of leukemic cells (bright Ig+, CD20+, CD22+, CD11+, CD25+, CD103+, CD123+).1,73 Immunohistochemical staining for Annexin A1 (ANXA1)74 and/or the search for the BRAF V600E mutation18,21,25,38 are very helpful in confirming the diagnosis.
Immunostaining for Annexin A1
ANXA1 was selected as a potential HCL diagnostic marker because at GEP it was found to be specifically upregulated in HCL.13 Immunostaining of BM biopsies with a monoclonal antibody directed against a fixative-resistant epitope of the ANXA1 protein (when run in parallel with an anti-CD20 monoclonal antibody) is highly specific for HCL at first diagnosis.74 However, it is not suitable for monitoring residual disease after therapy because the detection of low numbers of ANXA1+ HCL cells is made difficult by the presence of the overwhelming population of myeloid elements, macrophages, and T cells (that also express ANXA1).13,75-77 Under these circumstances, double immunostaining for ANXA1(cytoplasmic)/Pax5 (nuclear) or BRAF V600E mutation studies (see next section) may overcome the problem.
Detection of BRAF V600E by molecular assays and immunohistochemistry
The BRAF V600E mutation can be detected in BM or peripheral blood (PB) samples by Sanger sequencing or other more sensitive polymerase chain reaction (PCR)-based techniques18,21,25,38 (Figure 3). These molecular assays are usually applied to fresh samples but they may also work in archival Romanowsky-stained air-dried PB and BM smears and even in fixed-decalcified/paraffin-embedded BM trephine biopsies.78 An alternative approach for detecting BRAF V600E in routine paraffin sections is based on immunostaining with the monoclonal antibody (VE1) specifically directed against the BRAF V600E mutant.30,79 This antibody has been claimed to represent a reliable surrogate for molecular PCR assays80 but further studies are required to validate these findings
Differential diagnosis between HCL and its mimickers. (A) A needle biopsy of a bulky abdominal mass between the liver and the spleen, featuring mature lymphoid cells that infiltrate pancreatic glandular structures (left panel; hematoxylin and eosin [H&E] staining); the immunohistochemical staining with Annexin-1 (ANXA1; red labeling in the right panel) confirms the HCL origin of this infiltrate in such an unusual anatomic site. (B) A BM biopsy involved in SMZL with its typical intrasinusoidal infiltration pattern, which is highlighted by the B-cell markers CD79a (red; left panel) and PAX5 (brown; right panel). SMZL cells do not express ANXA1 (blue, right panel, with ANXA1-positive myeloid and T cells serving as internal control). Pictures of immunohistochemical stainings shown in panels A and B were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus). (C) A genetic diagnosis of HCL (vs HCL-v) can be easily made in whole-blood samples through BRAF-mutant allele-specific DNA PCR followed by agarose gel electrophoresis. The BRAF V600E gel band is visible in the blood sample from an HCL patient (known to contain <1% of leukemic cells by flow cytometry) but not in the blood sample from an HCL-v patient (containing >40% leukemic cells), which shows only the BRAF wild-type amplicon. The gel lanes to the left of the HCL and HCL-v samples represent a serial dilution (from 100% to 0%) of BRAF-mutant alleles with wild-type ones, which sets the detection limit of this qualitative assay at 0.05% mutant alleles. Dashed lines separate lanes of the same gels that were repositioned for clarity.
Differential diagnosis between HCL and its mimickers. (A) A needle biopsy of a bulky abdominal mass between the liver and the spleen, featuring mature lymphoid cells that infiltrate pancreatic glandular structures (left panel; hematoxylin and eosin [H&E] staining); the immunohistochemical staining with Annexin-1 (ANXA1; red labeling in the right panel) confirms the HCL origin of this infiltrate in such an unusual anatomic site. (B) A BM biopsy involved in SMZL with its typical intrasinusoidal infiltration pattern, which is highlighted by the B-cell markers CD79a (red; left panel) and PAX5 (brown; right panel). SMZL cells do not express ANXA1 (blue, right panel, with ANXA1-positive myeloid and T cells serving as internal control). Pictures of immunohistochemical stainings shown in panels A and B were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus). (C) A genetic diagnosis of HCL (vs HCL-v) can be easily made in whole-blood samples through BRAF-mutant allele-specific DNA PCR followed by agarose gel electrophoresis. The BRAF V600E gel band is visible in the blood sample from an HCL patient (known to contain <1% of leukemic cells by flow cytometry) but not in the blood sample from an HCL-v patient (containing >40% leukemic cells), which shows only the BRAF wild-type amplicon. The gel lanes to the left of the HCL and HCL-v samples represent a serial dilution (from 100% to 0%) of BRAF-mutant alleles with wild-type ones, which sets the detection limit of this qualitative assay at 0.05% mutant alleles. Dashed lines separate lanes of the same gels that were repositioned for clarity.
Differential diagnosis between HCL and HCL-like disorders
The immunohistochemical and molecular techniques may be particularly useful in confirming the diagnosis of HCL in unusual anatomical sites (Figure 3) and/or in distinguishing classic HCL from HCL-like disorders, such as splenic marginal zone lymphoma (SMZL), HCL variant (HCL-v),81 and splenic diffuse red pulp small B-cell lymphoma.81 This distinction is clinically important because classic HCL is treated differently from HCL-like disorders. The main features of HCL vs HCL-like disorders are briefly described in the following paragraphs and are summarized in Table 1.
Differential diagnosis between HCL and HCL-like neoplasms (SMZL, HCL-v, SDRPBCL)
| Features . | HCL . | SMZL . | HCL-v . | SDRPBCL . |
|---|---|---|---|---|
| Morphology | ||||
| Nucleus* | Oval, sometimes “coffee bean” shaped | Round | Round/oval | Round/oval, sometimes eccentric |
| Nucleolus* | Absent | Absent or small | Clearly evident | Absent or small |
| Cytoplasm* | Abundant, pale | Moderately abundant, basophil, sometimes with plasmacytoid features | Abundant | Moderately abundant, basophil, sometimes with plasmacytoid features |
| Cell surface | Thin, often long projections distributed all over the surface (circumferential) | Projections with a polar (noncircumferential) distribution | Similar to HCL | Short and thick projections with polar distribution |
| BM infiltration pattern | Diffuse and/or interstitial | Nodular and/or intrasinusoidal | Diffuse, intrasinusoidal, and/or interstitial | Intrasinusoidal, interstitial, and/or nodular |
| Splenic infiltration pattern | Red pulp, with effacement of white pulp | White pulp | Red pulp | Diffuse (red and white pulp) |
| Immunophenotype and genetic lesions | ||||
| Immunophenotypic markers | CD20+, CD5−, CD10−, CD23−CD103+, CD25+, CD11c+, CD123+, AnnexinA1+, BRAF V600E+ | CD20+, CD5−, CD10−, CD23−CD103−/+, CD25−/+, CD11c+/, CD123−, AnnexinA1−, BRAF V600E− | CD20+, CD5−, CD10−, CD23−CD103+, CD25−, CD11c+, CD123−, AnnexinA1−, BRAF V600E− | CD20+, CD5−/+, CD10−, CD23−/+,CD103−/+, CD25−, CD11c+, CD123−, AnnexinA1−, BRAF V600E− |
| Genetic lesions | BRAF V600E mutation in >97% of cases; KLF2 and CDKN1B mutations each in 16% of cases | Del(7q) in ∼30% of cases, NOTCH2 mutations in 10%-25% of cases, NF-κB pathway mutations in ∼35% of cases, KLF2 mutations in 20%-40% of cases | MAP2K1 mutations in 50% of cases; 17p (TP53) deletion in ∼30% of cases | Not well characterized |
| Features . | HCL . | SMZL . | HCL-v . | SDRPBCL . |
|---|---|---|---|---|
| Morphology | ||||
| Nucleus* | Oval, sometimes “coffee bean” shaped | Round | Round/oval | Round/oval, sometimes eccentric |
| Nucleolus* | Absent | Absent or small | Clearly evident | Absent or small |
| Cytoplasm* | Abundant, pale | Moderately abundant, basophil, sometimes with plasmacytoid features | Abundant | Moderately abundant, basophil, sometimes with plasmacytoid features |
| Cell surface | Thin, often long projections distributed all over the surface (circumferential) | Projections with a polar (noncircumferential) distribution | Similar to HCL | Short and thick projections with polar distribution |
| BM infiltration pattern | Diffuse and/or interstitial | Nodular and/or intrasinusoidal | Diffuse, intrasinusoidal, and/or interstitial | Intrasinusoidal, interstitial, and/or nodular |
| Splenic infiltration pattern | Red pulp, with effacement of white pulp | White pulp | Red pulp | Diffuse (red and white pulp) |
| Immunophenotype and genetic lesions | ||||
| Immunophenotypic markers | CD20+, CD5−, CD10−, CD23−CD103+, CD25+, CD11c+, CD123+, AnnexinA1+, BRAF V600E+ | CD20+, CD5−, CD10−, CD23−CD103−/+, CD25−/+, CD11c+/, CD123−, AnnexinA1−, BRAF V600E− | CD20+, CD5−, CD10−, CD23−CD103+, CD25−, CD11c+, CD123−, AnnexinA1−, BRAF V600E− | CD20+, CD5−/+, CD10−, CD23−/+,CD103−/+, CD25−, CD11c+, CD123−, AnnexinA1−, BRAF V600E− |
| Genetic lesions | BRAF V600E mutation in >97% of cases; KLF2 and CDKN1B mutations each in 16% of cases | Del(7q) in ∼30% of cases, NOTCH2 mutations in 10%-25% of cases, NF-κB pathway mutations in ∼35% of cases, KLF2 mutations in 20%-40% of cases | MAP2K1 mutations in 50% of cases; 17p (TP53) deletion in ∼30% of cases | Not well characterized |
In smears stained according to May-Grünwald Giemsa.
Unlike HCL, SMZL usually shows an intrasinusoidal infiltration pattern in the BM (clearly highlighted by immunohistochemistry for CD20) (Figure 3). Moreover, the SMZL cells circulating in the PB exhibit shorter hairy projections that are usually “polarized” to 1 area of the cell surface (unlike the circumferential hairy projections of HCL). Finally, SMZL cells are negative for the typical markers of HCL (especially CD103, CD25, and Annexin A1) (Figure 3), do not harbor BRAF V600E,21,38 and frequently carry Notch2 mutations.82,83
A more difficult task is the differential diagnosis between HCL and HCL-v. The most important criteria for the diagnosis of HCL-v are: (1) the higher frequency of leukocytosis (observed in only 10%-15% of HCL cases); (2) the absence of monocytopenia (a rather constant feature in HCL); (3) the morphological features of both HCL (hairy surface) and prolymphocytic leukemia (single prominent nucleolus); (4) the negativity for CD25, ANXA1, BRAF V600E, and CD123; and (5) the presence of MAP2K1 mutations84 in 50% of cases.
The splenic diffuse red pulp small B-cell lymphoma (SDRPBCL) shows several overlapping features with HCL-v because both tumors are negative for CD25, ANXA1, and BRAF V600E.81,85 The red pulp is infiltrated by tumor cells with plasmacytoid appearance. Genomic studies have not been performed in SDRPBCL.
Therapeutic implications of BRAF V600E mutation in HCL
Although a rare disease, during the past 50 years, HCL has served as a paradigm for the development of new forms of therapy. For >2 decades, the only available therapeutic intervention in HCL was splenectomy. The first significant improvement in the care of this disease occurred in the early 80s, with the introduction of interferon.86 This drug usually induced a partial response, sometimes durable, in 40% to 80% of patients.87,88 Currently, the gold-standard therapy for HCL is based on the purine analogs cladribine and pentostatin89,90 (introduced in the 90s), both of which induce durable complete responses (CRs) in 80% to 85% of patients.3,91 Combining cladribine with the anti-CD20 monoclonal antibody (rituximab) may produce even better results.92
In spite of these great advances, disease relapses occur in ∼40% to 50% of patients and these events are associated with a progressively worse response to purine analogs.93,94 Moreover, over time, therapy with purine analogs can lead to cumulative myelotoxic effects and profound immune suppression. Anti-CD22 immunotoxin (moxetumomab pasudotox) showed high clinical activity in this setting of patients.95 More recently, the availability of BRAF and MEK inhibitors, originally developed for the treatment of BRAF-mutated metastatic melanoma,96,97 offer a new therapeutic opportunity for HCL patients with refractory or relapsed disease.
Preclinical studies with BRAF inhibitors
Preclinical studies in HCL have been difficult to perform due to the absence of true human HCL cell lines5,6 and animal models of the disease.7 Therefore, the activity of inhibitors of BRAF (vemurafenib and dabrafenib) and MEK (trametinib) has been investigated in vitro on primary leukemic cells purified from HCL patients,52 a rather difficult task because of the BM fibrosis and the low percentage of circulating tumor cells that are typical of HCL. Notably, BRAF or MEK inhibitors caused marked dephosphorylation of MEK/ERK in HCL cells, silencing of the RAS-RAF-MEK-ERK pathway transcriptional output, loss of the HCL-specific GEP signature, change of the morphology from “hairy” to “smooth,” and eventually apoptosis.52 Usually, apoptosis followed changes in hairy morphology (Figure 2) and was associated with the upregulation of some proapoptotic genes, such as BIM/BCL2L11 and CDKN1C/p57-KIP2.52 These findings are in keeping with the concept that HCL growth is predominantly sustained by inhibition of apoptosis52 and that the proliferative index of HCL (<5%) is 1 of the lowest among B-cell neoplasms.58 Interestingly, BRAF inhibitors induced these effects specifically in HCL but not in primary leukemic cells from patients with HCL-like disorders. Collectively, these results strongly supported and informed the clinical use of BRAF and MEK inhibitors in HCL.
Therapy of relapsed/refractory HCL with BRAF inhibitors
The clinical activity of BRAF inhibitors was initially described in anecdotal cases of refractory/relapsed HCL.98-105 Recently, we reported the results of 2 phase-2 clinical trials (1 carried out in Italy and the other in the United States) in 54 patients with refractory/relapsed HCL treated with the BRAF inhibitor vemurafenib.63 In both studies, the drug was administered at the dose of 960 mg twice daily (as previously used in metastatic melanoma) for a median of 16 weeks in the Italian study and 18 weeks in the US study.63 A total of 49 patients were available for response. The overall response rates were 96% (CR = 35%; Figure 4) and 100% (CR = 42%) in the Italian trial (n = 25 patients) and the US study (n = 24 patients), respectively. Response to the drug was rapid, usually occurring within a median of 2 to 3 months.63
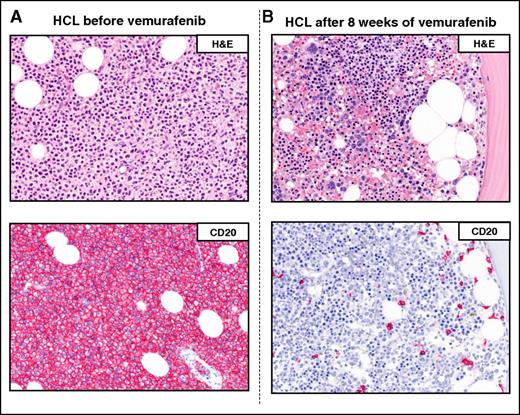
Example of a complete remission obtained with vemurafenib in relapsed/refractory HCL. The BM biopsies of a patient with multiple HCL relapses is shown (A) before and (B) after 8 weeks of treatment with vemurafenib. H&E (top panels) and CD20 (red; bottom panels) stainings show a massive leukemic infiltration which is largely cleared by vemurafenib (<10% residual HCL cells posttherapy). Pictures of immunohistochemical stainings were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus).
Example of a complete remission obtained with vemurafenib in relapsed/refractory HCL. The BM biopsies of a patient with multiple HCL relapses is shown (A) before and (B) after 8 weeks of treatment with vemurafenib. H&E (top panels) and CD20 (red; bottom panels) stainings show a massive leukemic infiltration which is largely cleared by vemurafenib (<10% residual HCL cells posttherapy). Pictures of immunohistochemical stainings were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus).
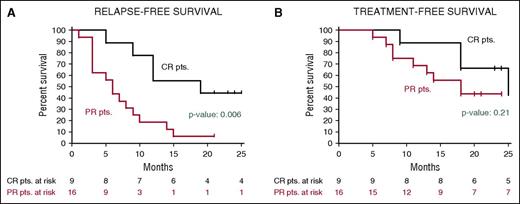
After a median follow-up of 23 months, the median relapse-free survival in the Italian trial was 19 months in patients who had achieved a CR and 6 months in those who had obtained a partial response63 (Figure 5); the median treatment-free survival in these cases was 25 and 18 months, respectively. In the US study, the progression-free survival and the overall survival at 1 year were 73% and 91%, respectively.63
Relapse-free and treatment-free survival after vemurafenib in relapsed/refractory HCL. Kaplan-Meier curves displaying the survival free (A) from relapse and (B) from a subsequent antileukemic treatment in the HCL-PG01 phase-2 Italian trial at a median follow-up of 23 months after the last vemurafenib dose. Relapse was defined as the reappearance of 1 or more cytopenias (neutrophils, <1.5 × 109/L; platelets, <100 × 109/L; or hemoglobin, <11 g/dL). Time to relapse (A) was longer (by log-rank test) in patients who achieved a complete remission vs a partial remission. This difference was less evident regarding time to next treatment (B), as various patients relapsing after a partial remission developed mild cytopenias (eg, ∼1 N × 109/L or ∼80 platelets × 109/L) which remained relatively stable for some time and did not immediately trigger a subsequent treatment. Reprinted from Tiacci et al63 with permission.
Relapse-free and treatment-free survival after vemurafenib in relapsed/refractory HCL. Kaplan-Meier curves displaying the survival free (A) from relapse and (B) from a subsequent antileukemic treatment in the HCL-PG01 phase-2 Italian trial at a median follow-up of 23 months after the last vemurafenib dose. Relapse was defined as the reappearance of 1 or more cytopenias (neutrophils, <1.5 × 109/L; platelets, <100 × 109/L; or hemoglobin, <11 g/dL). Time to relapse (A) was longer (by log-rank test) in patients who achieved a complete remission vs a partial remission. This difference was less evident regarding time to next treatment (B), as various patients relapsing after a partial remission developed mild cytopenias (eg, ∼1 N × 109/L or ∼80 platelets × 109/L) which remained relatively stable for some time and did not immediately trigger a subsequent treatment. Reprinted from Tiacci et al63 with permission.
In both trials, the toxicity of vemurafenib was similar to that previously reported in patients with metastatic melanoma.106-108 Drug-related adverse events mostly affected the skin (especially rash, photosensitivity, palmar/plantar fibrosis, warts) and the joints (arthralgia, arthritis). Dose reduction was required in 50% to 58% of patients in the 2 trials.63 Patients with arthralgias may benefit from a low dose of steroids.
Secondary tumors were observed in a total of 7 patients. Among the 3 patients in the Italian trial, 2 developed a basal cell carcinoma and 1 a superficial melanoma; 3 and 1 patients from the US study developed squamous cell carcinoma and a basal cell carcinoma, respectively.63 All tumors were managed by simple excision. Similar to what was observed in patients with metastatic melanoma, these tumors are likely related to a paradoxic effect of the BRAF inhibitor toward the skin keratinocytes or melanocytes when they carry a preexisting RAS mutation (especially affecting H-RAS).109
Notably, vemurafenib showed no significant myelotoxicity. This suggest its potential usefulness not only in refractory/relapsed HCL patients but also as a frontline drug in patients who have to discontinue purine analogs because of severe side effects or in patients who, at the time of initial diagnosis, present with a severe opportunistic infection that precludes the use of myelotoxic agents.110
Mechanisms of resistance to BRAF inhibitors in HCL
Similar to what was observed in metastatic melanoma,111-113 the dramatic responses achieved with vemurafenib in refractory/relapsed HCL were frequently followed by relapse, likely due to development of mechanisms of resistance to the BRAF inhibitor. At least in melanoma, the resistance mechanisms so-far identified in vitro and also confirmed in patients include: (1) the activation of tyrosine kinase receptors (eg, MET114 ) via interaction with microenvironmental factors (eg, HGF114 ) leading to the reactivation of the RAS-RAF-MEK-ERK pathway through CRAF, and also to the activation of alternative signaling pathways, such as PI3K-AKT; (2) the emergence of N-RAS115 mutations that induce the reactivation of the RAS-RAF-MEK-ERK signaling pathway, mainly through CRAF116 ; (3) the emergence of activating mutations of MEK117 ; (4) the overexpression of COT (encoded by the MAP3K8 gene),118 a protein kinase that can activate ERK through a MEK-dependent (but BRAF-independent) mechanism; (5) the copy-number amplification of the BRAF mutant119 ; and (6) the alternative splicing of the BRAF mutant leading to the generation of a smaller molecule able to form dimers that can evade the action of BRAF inhibitors.120
At present, there is only scarce knowledge of the mechanisms of resistance to BRAF inhibitors operating in HCL. Like in melanoma, acquired mutations of BRAF are not implicated in the mechanism of resistance. In at least 1 patient in the phase-2 HCL trial, resistance to vemurafenib was due to the emergence of K-RAS mutations.63 Interestingly, at immunohistochemistry in the BM biopsy, 6 of 13 patients in the Italian trial showed residual leukemic cells that still expressed phospho-ERK despite prolonged exposure (16-20 weeks) to vemurafenib. These findings strongly suggest that, in at least some HCL patients, the growth of leukemic cells is still closely dependent on the RAS-RAF-MEK-ERK signaling pathway that is likely reactivated through mechanisms that bypass the activity of the BRAF inhibitors. Functional studies and gene sequencing of leukemic cells before and after treatment with BRAF inhibitors are needed to further elucidate the mechanisms of resistance to these agents in HCL.
Future perspectives
Due to the lack of true HCL cell lines5,6 and the paucity of primary HCL cells that can usually be collected from HCL patients, a faithful BRAF V600E–based mouse model of the disease could be extremely useful both for functional studies and for testing the activity of new drugs. Moreover, because BRAF inhibitors do not appear to fully eradicate the disease, further studies on the nature of preleukemic HSCs7 may have pathogenetic implications.
In anecdotal cases, lower doses of vemurafenib (eg, 240 mg twice daily) proved to be effective in HCL patients with refractory or relapsed disease.98-100,121,122 In the phase-2 HCL trials,63 vemurafenib was administered at the dose of 960 mg twice daily (as previously used in metastatic melanoma) and mostly reduced to 720 mg (in the Italian trial) or 480 mg (in the US trial), sometimes followed by reescalation. Prospective clinical studies are necessary to investigate the optimal dose and treatment duration of BRAF inhibitors in HCL.
The duration of the response to BRAF inhibitors in refractory/relapsed HCL patients appears to correlate with the depth of the response (being usually longer in patients achieving a CR).63 It has been reported that duration of response to BRAF inhibitors in naive HCL patients may be shorter than that achievable with purine analogs.122 However, these results are based on anecdotal cases treated with a low dose of vemurafenib.
Future therapeutic strategies should be directed to overcome the mechanism of resistance with the aim of improving the rate and duration of CR and possibly reducing the incidence of secondary skin tumors. Based on previous experience in melanoma patients, clinical trials using dabrafenib (another BRAF inhibitor) alone or BRAF plus MEK inhibitors for dual targeting of the RAS-RAF-MEK-ERK signaling pathway97 are also ongoing in refractory/relapsed HCL. Combining a BRAF inhibitor with an anti-CD20 monoclonal antibody represents another interesting therapeutic strategy. The rationale for this approach is to use immunotherapy to eradicate the residual BRAF inhibitor-resistant HCL cells.
Acknowledgments
The authors thank L. De Carolis, G. Schiavoni, V. Pettirossi, A. Santi, and P. Sportoletti for their contribution to their research program on hairy cell leukemia.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (MCO-10007 [B.F.] and IG-14447) (E.T.), the European Research Council (FP7/2007-2013 “Hairy Cell Leukemia,” 617471) (E.T.), the Hairy Cell Leukemia Foundation (B.F. and E.T.), Ministero dell’Istruzione, Università e Ricerca (PRIN 20104HBZ8E [B.F.] and Futuro in Ricerca 2010-RBFR10WT2K [E.T.]), Ministero della Salute (Giovani Ricercatori GR-2010-2303444) (E.T.), the Associazione Umbra contro le Leucemie e i Linfomi (B.F.), and Roche Pharmaceuticals (B.F.). E.T. is a Scholar in Clinical Research of the Leukemia & Lymphoma Society (contract no. 2030-14).
Authorship
Contribution: B.F. wrote the text and prepared the table; E.T. prepared the figures and helped in writing the text and preparing the table; and M.P.M. commented on, edited, and approved the paper.
Conflict-of-interest disclosure: B.F. and E.T. filed a patent on the use of the BRAF V600E mutation as an HCL biomarker. B.F. received research funding from Roche. M.P.M. declares no competing financial interests.
Correspondence: Brunangelo Falini, Institute of Hematology and Center for Hemato-Oncology Research (C.R.E.O.), University and Hospital of Perugia, Perugia, Italy; e-mail: brunangelo.falini@unipg.it.

![Figure 3. Differential diagnosis between HCL and its mimickers. (A) A needle biopsy of a bulky abdominal mass between the liver and the spleen, featuring mature lymphoid cells that infiltrate pancreatic glandular structures (left panel; hematoxylin and eosin [H&E] staining); the immunohistochemical staining with Annexin-1 (ANXA1; red labeling in the right panel) confirms the HCL origin of this infiltrate in such an unusual anatomic site. (B) A BM biopsy involved in SMZL with its typical intrasinusoidal infiltration pattern, which is highlighted by the B-cell markers CD79a (red; left panel) and PAX5 (brown; right panel). SMZL cells do not express ANXA1 (blue, right panel, with ANXA1-positive myeloid and T cells serving as internal control). Pictures of immunohistochemical stainings shown in panels A and B were taken with the 40×/0.85 objective (U Plan Apo) of a BX61 microscope equipped with a DP71 digital camera, using cell^B x.y acquisition software (all from Olympus). (C) A genetic diagnosis of HCL (vs HCL-v) can be easily made in whole-blood samples through BRAF-mutant allele-specific DNA PCR followed by agarose gel electrophoresis. The BRAF V600E gel band is visible in the blood sample from an HCL patient (known to contain <1% of leukemic cells by flow cytometry) but not in the blood sample from an HCL-v patient (containing >40% leukemic cells), which shows only the BRAF wild-type amplicon. The gel lanes to the left of the HCL and HCL-v samples represent a serial dilution (from 100% to 0%) of BRAF-mutant alleles with wild-type ones, which sets the detection limit of this qualitative assay at 0.05% mutant alleles. Dashed lines separate lanes of the same gels that were repositioned for clarity.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/15/10.1182_blood-2016-07-418434/4/m_1918f3.jpeg?Expires=1769847845&Signature=u2LWkwjEip2IFXX53hPlaOYtjOSZ9Q6absaKHlHVA-VVJ93qpSVHZXH9sOzkzIoTOB8NoP7ltuP9tFKZB7McJPOhRdH648oeS1qYpS0BacCKNh-ELFT3fheUonl5oNEg5kbaBQuGv2nOUFMZCFY09ssuGUxTjrMDfMyX~0CPKUWvcKu~J1QcCNpSjeqaaCfc1Bk6WJZ7kF-SidNEMBlQKd4fWHexyZPs5WzJgKxix-N~OeMjkmZtWXfRq4X0S6MQ-Y~CFIMrl-4xqZ5NvOYqclb8tI30WkJs3VE8CRulWP3uWVTOe7GmxH5BfLzVpLynUXSiQsJ7VvzlvKbxqBIwXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)