Key Points
Using AML as a model, we investigated the effect of treatment and disease evolution on functionally defined cancer stem cell populations.
We demonstrate large-scale changes in LSC frequency and phenotype after relapse, best described using high-dimensional space analyses.
Abstract
Most cancers evolve over time as patients initially responsive to therapy acquire resistance to the same drugs at relapse. Cancer stem cells have been postulated to represent a therapy-refractory reservoir for relapse, but formal proof of this model is lacking. We prospectively characterized leukemia stem cell populations (LSCs) from a well-defined cohort of patients with acute myelogenous leukemia (AML) at diagnosis and relapse to assess the effect of the disease course on these critical populations. Leukemic samples were collected from patients with newly diagnosed AML before therapy and after relapse, and LSC frequency was assessed by limiting dilution analyses. LSC populations were identified using fluorescent-labeled cell sorting and transplantation into immunodeficient NOD/SCID/interleukin 2 receptor γ chain null mice. The surface antigen expression profiles of pretherapy and postrelapse LSCs were determined for published LSC markers. We demonstrate a 9- to 90-fold increase in LSC frequency between diagnosis and relapse. LSC activity at relapse was identified in populations of leukemic blasts that did not demonstrate this activity before treatment and relapse. In addition, we describe genetic instability and exceptional phenotypic changes that accompany the evolution of these new LSC populations. This study is the first to characterize the evolution of LSCs in vivo after chemotherapy, identifying a dramatic change in the physiology of primitive AML cells when the disease progresses. Taken together, these findings provide a new frame of reference by which to evaluate candidate AML therapies in which both disease control and the induction of more advanced forms of disease should be considered.
Introduction
The cancer stem cell model proposes that rare populations of malignant cells are responsible for maintaining the bulk tumor and are believed to represent therapy-refractory reservoirs for relapse. Cancer stem cell populations (CSCs) have been proposed for chronic myelogenous leukemia (CML) and acute myelogenous leukemia (AML), as well as most solid organ cancers. Substantial heterogeneity of CSC phenotypes has been reported, and it is not clear that all cancers possess a consistently identifiable human CSC phenotype.1,2 AML was the first malignancy for which a prospectively identifiable CSC population was described and has remained a model for the study of CSCs. In AML, isolation of leukemic stem cells, LSCs, has historically been based on the surface expression of CD34 and CD38,3 as initial publications supported restriction of LSC activity to the CD34+CD38− population.4,5 After the initial publication by Lapidot et al in 1994,4 additional markers of LSCs have been proposed, including CD123,6,7 CD32,8 CD33,9 CD45RA,10 CD47,11 CD96,12 CD99,13 IL1RAP,14 and TIM-3.15 Recent studies have identified increasing intrapatient and interpatient heterogeneity of AML LSC populations.16-19 A direct assessment of intrapatient evolution of LSCs that may occur during disease pathogenesis and/or treatment has not been reported.
We investigated the frequency and surface antigen phenotypes of functionally defined LSCs in paired primary human AML samples at diagnosis and in relapse. Surprisingly, we found that LSC frequencies and phenotypic diversity are much greater at relapse than before initial therapy. Our data indicate that current AML therapeutic regimens may promote dramatic changes in the LSC compartment. A better understanding of how these changes occur will aid in the development of novel therapeutics for patients with relapsed disease.
Materials and methods
Patients
Twenty-five patients with AML who relapsed after achieving a morphologic remission after induction therapy were selected for this study (supplemental Table 1, available on the Blood Web site). Bone marrow or peripheral blood samples were obtained after informed consent at diagnosis and relapse under institutional review board–approved protocols. Bone marrow aspirates from healthy donors (NBM) were obtained after informed consent of an institutional review board–approved protocol. All samples were processed as previously described.20,21
Immunophenotyping/fluorescence-activated cell sorting
Thawed primary AML samples at diagnosis and relapse were stained with mouse anti-human antibodies against CD34 (phycoerythrin-Cy-7 conjugate, 8G12; BD BioSciences), CD38 (allophycocyanin [APC], HB7 [BD BioSciences] or peridinin chlorophyll protein complex–Cy5.5 conjugate, HIT2 [BD Pharmingen]), and leukemia-specific aberrant markers including CD32 (APC, 6C4; eBioScience), CD33 (fluorescein isothiocyanate, HIM3-4; BD Pharmingen), CD45RA (APC-Cy7, HI100; BioLegend), CD47 (APC, B6H12; eBioScience), CD96 (PE, NK92.39; eBioScience), CD97 (fluorescein isothiocyanate, VIM3b; BD Pharmingen), CD99 (PE, 3B2/TA8; eBioScience), CD123 (PE, 7G3; BD Pharmingen), HLA-DR (APC-Cy7, L243; BD BioSciences), IL1RAP (PE, 89412; R&D Systems), and TIM-3 (PE, F38-2E2; BioLegend). DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) was used to exclude nonviable cells. Flow cytometry was performed on a LSR II (BD Biosciences), and data were analyzed using FlowJo (TreeStar, Ashland, OR). For sorting, paired primary AML samples at diagnosis and relapse were stained using antibodies against CD3 (PE-Cy5, HIT3a; BioLegend), CD32, CD34, and CD38 with DAPI in PBS plus 0.5% HI-FBS, followed by sorting, using a FACSAria cell sorter (BD Biosystems). To prevent the development of graft vs host disease in recipient mice, T-cell depletion was performed by excluding CD3+ cells. The purity of each sort was subsequently assessed and verified.
Xenotransplantation/limiting dilution analysis
All animal studies were performed in compliance with the University of Rochester Medical Center Laboratory Animal Care regulations on protocol UCAR-2012-026. NOD/SCID/interleukin 2 receptor γ chain null (NOD/SCID/IL2rγ−/−, termed NSG) recipient mice received a dose of radiation (250 cGy) using a RadSource X-ray irradiator 24 hours before transplantation. Two doses of human intravenous immune globulin were administrated (0.5 mg/g) by intraperitoneal injection on the day of irradiation, as well as the day of transplantation. For samples with fewer than 1 million human cells per animal, syngeneic donor (NSG) splenocytes were mixed with the primary AML sample before transplantation. Primary bulk AML cells were serially diluted (cell dose range, 1 × 103 to 5 × 106) and transplanted into NSG mice via the tail-vein route. Engraftment was assessed 12 weeks posttransplantation, or earlier if recipients appeared moribund. The frequency of LSCs was calculated with the number of positively engrafted mice at each dose (L-Calc Software; STEMCELL Technologies). For functional assessment of LSC activity on sorted populations, sorted cells were injected into mice via tail-vein injection (final volume of 0.2 mL). The number of cells transplanted for each sorted population was proportional to the contribution of that population to the patient’s original sample. The equivalent dose of mononuclear cells per mouse was from 5 × 105 to 3 × 106 cells for sorted populations. An unsorted control group was injected with 1 × 106 bulk cells per mouse. Mice were killed at 12 weeks posttransplantation or any time they appeared moribund. Recipient bone marrow (BM) was analyzed by flow cytometry, using anti-mouse CD45 (APC, 30-F11; BD Pharmingen), anti-human CD45 (PE-Cy5, HI30; BioLegend), anti-human CD19 (PE, HIB19; BD Pharmingen), and CD97 (fluorescein isothiocyanate, VIM3b; BD Pharmingen) to verify engraftment. Engraftment was defined as the presence of more than 0.1% live human CD45+ cells in the sample. Engraftment with more than 50% of human CD45+ events positive for CD19 and lacking CD97 expression were classified as having normal engraftment. The expression of other antigens (CD32, CD33, CD45RA, CD47, CD96, CD123, HLA-DR, and TIM-3) was also performed to confirm the leukemic engraftment.
Statistics
Immunophenotyping data were analyzed for their relevance, using 2-tailed Mann-Whitney test (NBM vs AML) or Wilcoxon matched-pairs signed rank test (diagnosis vs relapse). A P value < .05 was considered significant.
Results
LSC frequency after relapse
To directly assess the effect of chemotherapy and relapse on LSC frequency, we performed limiting dilution analyses (LDA) on unsorted cells from 5 patients at diagnosis and at relapse (Table 1; supplemental Table 2). Decreasing doses of leukemic cells are transplanted into NSG recipients to determine the fewest number of cells required for human leukemic engraftment. Similar to previously published results, we confirmed that LSCs are rare with considerable interpatient heterogeneity in LSC frequencies (range, 1 in 9 × 103 to 1 in 2.3 × 106 cells). At relapse, LSCs still represent a minor fraction of the leukemic mass, with frequencies varying between 1 in < 1 × 103 and 1 in 1 × 105 cells in our limited cohort. Direct comparison of pretherapy and relapse samples for individual patients demonstrated a previously unreported 9- to 90-fold increase in LSC frequency (Table 1; supplemental Table 2) after disease relapse. These results show for the first time a significant increase in LSC pool size after relapse in patients with AML.
LSC frequency analysis for paired diagnosis and relapse samples.
| AML . | Status . | Cells injected, n . | Frequency . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1E3 . | 5E3 . | 1E4 . | 5E4 . | 1E5 . | 5E5 . | 1E6 . | 5E6 . | |||
| AML1 | Diagnosis | — | — | — | 0/4 | 0/3 | 3/4 | 3/3 | 1/1 | 428 455 |
| AML1 | Relapse | — | — | 3/3 | 5/5 | 5/5 | 5/5 | 5/5 | 4/4 | <10 000 |
| AML3 | Diagnosis | — | 3/5 | 2/4 | 5/5 | 5/5 | 5/5 | 3/3 | — | 8 815 |
| AML3 | Relapse | 4/4 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 2/2 | — | <1 000 |
| AML15 | Diagnosis | — | — | 0/5 | 0/5 | 1/5 | 1/5 | 1/2 | 4/5 | 2 297 888 |
| AML15 | Relapse | 0/5 | 1/5 | 1/5 | 1/4 | 3/5 | 5/5 | 2/2 | — | 92 862 |
| AML18 | Diagnosis | — | 1/5 | 1/4 | 3/5 | 2/3 | 4/4 | 1/1 | — | 56 116 |
| AML18 | Relapse | 4/5 | 5/5 | 3/3 | 4/4 | 4/4 | — | — | — | 619 |
| AML22 | Diagnosis | — | 0/2 | 1/3 | 1/5 | 2/4 | 2/4 | 1/2 | 1/1 | 472 106 |
| AML22 | Relapse | 0/3 | 1/3 | 4/5 | 2/2 | 2/4 | 3/3 | 2/2 | — | 38 300 |
| AML . | Status . | Cells injected, n . | Frequency . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1E3 . | 5E3 . | 1E4 . | 5E4 . | 1E5 . | 5E5 . | 1E6 . | 5E6 . | |||
| AML1 | Diagnosis | — | — | — | 0/4 | 0/3 | 3/4 | 3/3 | 1/1 | 428 455 |
| AML1 | Relapse | — | — | 3/3 | 5/5 | 5/5 | 5/5 | 5/5 | 4/4 | <10 000 |
| AML3 | Diagnosis | — | 3/5 | 2/4 | 5/5 | 5/5 | 5/5 | 3/3 | — | 8 815 |
| AML3 | Relapse | 4/4 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 2/2 | — | <1 000 |
| AML15 | Diagnosis | — | — | 0/5 | 0/5 | 1/5 | 1/5 | 1/2 | 4/5 | 2 297 888 |
| AML15 | Relapse | 0/5 | 1/5 | 1/5 | 1/4 | 3/5 | 5/5 | 2/2 | — | 92 862 |
| AML18 | Diagnosis | — | 1/5 | 1/4 | 3/5 | 2/3 | 4/4 | 1/1 | — | 56 116 |
| AML18 | Relapse | 4/5 | 5/5 | 3/3 | 4/4 | 4/4 | — | — | — | 619 |
| AML22 | Diagnosis | — | 0/2 | 1/3 | 1/5 | 2/4 | 2/4 | 1/2 | 1/1 | 472 106 |
| AML22 | Relapse | 0/3 | 1/3 | 4/5 | 2/2 | 2/4 | 3/3 | 2/2 | — | 38 300 |
LDA was performed by injecting increasing numbers of unsorted unfractionated leukemic marrow cells from 5 paired primary patient samples at diagnosis and in relapse into recipient mice (range, 1000-5 000 000). The number of mice demonstrating the presence of human leukemia in their marrow at time of harvest (numerator), as well as the number of mice transplanted (denominator), is indicated for each cell dose for each sample. LSC frequency, calculated using L-Calc Software (STEMCELL Technologies), is reported, as well as the upper and lower estimates for each sample.
1E3, 1000; 5E3, 5000; 1E4, 10 000, 5E4, 50 000; 1E5, 100 000; 5E5, 500 000; 1E6, 1 000 000; 5E6, 5 000 000.
LSC phenotype diversifies after relapse
To assess the effect of chemotherapy and relapse on the surface antigen phenotype of the LSC populations within individual patients, we analyzed the expression of CD34 and CD38, the 2 most commonly employed markers for LSC activity (supplemental Figures 1 and 2), on normal bone marrow samples, as well as paired diagnosis and relapse AML samples. A subset of patients with AML do not express CD34 on their blasts; for these samples, we employed CD32, a transmembrane glycoprotein expressed on B cells, granulocytes, monocytes, and macrophages (supplemental Figure 3).22 CD32 expression has been shown to be associated with LSC activity in some patients with AML,8 and in combination with CD38, CD32 is capable of segregating AML samples into 4 distinct populations. Despite the marked increase in LSC frequency at relapse, the surface expression of CD32, CD34, and CD38 remained generally stable between diagnosis and relapse, as assessed by comparative studies on 25 paired specimens (supplemental Figures 1-3).
To functionally assess the effect of chemotherapy and relapse on the surface antigen phenotype of the patients’ specific LSC populations, we identified 7 of the 25 patients’ prechemotherapy samples capable of establishing leukemic engraftment in NSG mice with sufficient material to perform a detailed analysis of LSC activity. Four individual populations from each patient, based on the expression of CD34 and CD38 or CD32 and CD38 expression, were sorted and individually assessed for LSC activity, using the NSG model (Figure 1; supplemental Figure 4). This model was selected because it has been shown to be a robust model for studying AML, offering high-level engraftment for most specimens and a lack of residual innate immunity, which has been shown to bias experimental results.16,23 Postsort, the number of cells transplanted for each population was proportional to the contribution of that population to the patient’s original sample (supplemental Table 3). We chose this strategy because injecting an equal number of cells for each sorted population does not reflect the actual contribution of the population to the patient’s disease and may over- or underestimate the LSC potential of a given population.
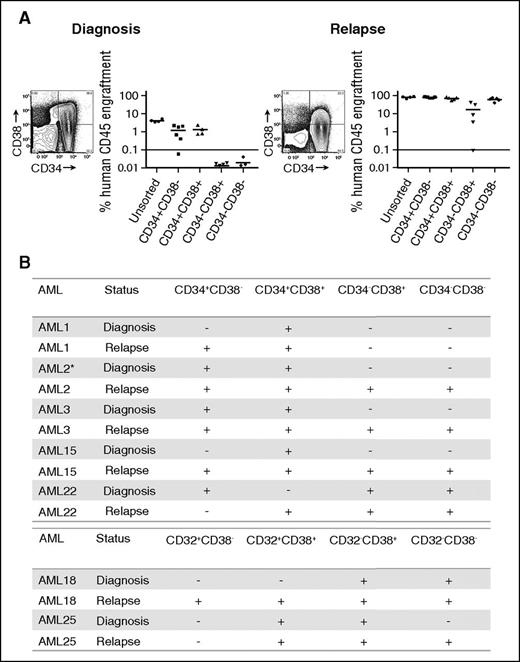
LSC populations from paired AML diagnosis and relapse samples. (A) Engraftment levels of sorted cells of 4 populations (CD34+CD38−, CD34+CD38+, CD34−CD38+, CD34−CD38−), as well as unsorted cells from a representative AML sample (AML3) at diagnosis and relapse. Each dot represents the human CD45+ engraftment level in BM of individual NSG mouse; a percentage greater than 0.1 is defined as leukemic engraftment. The horizontal line is the mean level of engraftment for each population. (B) Summary of the LSC activity in 7 sorted AML samples (as defined by CD34 and CD38 or CD32 and CD38) at diagnosis and relapse. The equivalent dose of mononuclear cells per mouse is from 5 × 105 to 3 × 106 cells for sorted populations. The summary of cell dose for primary xenotransplantation is in supplemental Table 3. Plus sign indicates LSC detected at this sensitivity; minus sign indicates no LSC detected at this sensitivity. *LSC activity detected using NOD-scid IL2Rgnull-3/GM/SF mice because of limited materials.
LSC populations from paired AML diagnosis and relapse samples. (A) Engraftment levels of sorted cells of 4 populations (CD34+CD38−, CD34+CD38+, CD34−CD38+, CD34−CD38−), as well as unsorted cells from a representative AML sample (AML3) at diagnosis and relapse. Each dot represents the human CD45+ engraftment level in BM of individual NSG mouse; a percentage greater than 0.1 is defined as leukemic engraftment. The horizontal line is the mean level of engraftment for each population. (B) Summary of the LSC activity in 7 sorted AML samples (as defined by CD34 and CD38 or CD32 and CD38) at diagnosis and relapse. The equivalent dose of mononuclear cells per mouse is from 5 × 105 to 3 × 106 cells for sorted populations. The summary of cell dose for primary xenotransplantation is in supplemental Table 3. Plus sign indicates LSC detected at this sensitivity; minus sign indicates no LSC detected at this sensitivity. *LSC activity detected using NOD-scid IL2Rgnull-3/GM/SF mice because of limited materials.
Before therapy, we observed LSC activity not only within CD34+CD38− populations but also within additional populations, confirming previous reports that the LSC phenotype demonstrates considerable intra- and interpatient heterogeneity.17-19 Most patients demonstrated LSC activity in more than 1 population at diagnosis, and CD32, CD34, or CD38 expression were not universally associated with LSC activity.
Examining LSC activity in the paired relapse samples from the same patients revealed LSC activity in immunophenotypically defined populations of leukemic cells in which LSC activity was not detected in the prechemotherapy sample (Figure 1; supplemental Figure 4) regardless of the antigen pair employed. In all but 1 patient (AML22), the number of phenotypically distinct LSC populations (as defined by CD34 and CD38 or CD32 and CD38) increased after relapse. For example, in AML3, LSC ability was detected in CD34+CD38+ and CD34+CD38− populations at diagnosis. At relapse, LSC ability was maintained in the CD34+CD38+ and CD34+CD38− populations but was also present in the CD34−CD38+ and CD34−CD38− populations (Figure 1A). We confirmed that LSC populations fulfilled the criteria required for cancer stem cell populations24 by performing secondary transplantation studies (supplemental Table 4) and assessing their ability to recapitulate the patients’ original surface antigen phenotype (supplemental Figure 5). The majority of populations demonstrating engraftment in primary recipients also engrafted secondary recipient mice. The most common reason for failure to engraft secondary recipient mice was insufficient cell dose from the primary recipient marrow. Overall, these data demonstrate that increase in LSC frequency identified in our LDA is distributed across a greater number of phenotypically distinct populations at relapse.
Known leukemia stem cell antigens do not correlate with increased LSC pool at relapse
Our analysis of pretherapy and postrelapse AML samples demonstrated the relative stability of CD34- and CD38-defined populations between time of diagnosis and relapse (supplemental Figures 1 and 2). Since the initial report by Dick and Lapidot,4 additional surface antigens have been shown to be capable of enriching for LSC populations from some patients with AML.6,8-15,25,26 To assess whether the increase in the LSC pool size at relapse correlated with the frequency of populations defined by published LSC markers, we determined the frequency of populations expressing CD32, CD33, CD45RA, CD47, CD96, CD97, CD123, HLA-DR, TIM-3, CD99, and IL1RAP before chemotherapy and after relapse (Figures 2 and 3; supplemental Figure 6). Representative data from the CD123 and TIM-3 analyses are shown in Figure 2B and indicate no substantial increase in either marker at relapse, despite the large increase in LSC frequency noted in Table 1. Further analyses of CD33, CD97, CD99, TIM-3, and HLA-DR expression (summarized in Figure 3) indicate that although each marker readily distinguishes leukemic blasts from normal BM cells, there is no consistent change between diagnosis and relapse AML that correlates with the large increase in LSC frequency at relapse. Similarly, CD123, CD97, IL1RAP, TIM-3, and CD45RA readily distinguished leukemic CD34+CD38- cells and CD34+CD38+ cells from their normal counterparts. Of all the marker and combinations examined, only the frequency of the CD123+CD34+CD38− population increased between diagnosis and relapse (67.6% ± 28.5% vs 84.2% ± 17.5%; P = .001).
Expression profiles of LSC markers on paired AML diagnosis and relapse samples. (A) A representative example of LSC marker immunophenotyping strategy in AML bulk cells (AML3, diagnosis). The LSC panel included CD32, CD33, CD45RA, CD47, CD96, CD97, CD99, CD123, HLA-DR, IL1RAP, and TIM-3 (top). (B) Frequencies of LSC marker positive populations (CD123+ and TIM-3+) in bulk AML cells at diagnosis (middle) and relapse (right) compared with NBM cells (left) (NBM, n = 22-23; AML, n = 24-25). Mean ± standard error of the mean values are plotted. *P < .05; **P < .01; ***P < .001; ****P < .0001. NS, not significant.
Expression profiles of LSC markers on paired AML diagnosis and relapse samples. (A) A representative example of LSC marker immunophenotyping strategy in AML bulk cells (AML3, diagnosis). The LSC panel included CD32, CD33, CD45RA, CD47, CD96, CD97, CD99, CD123, HLA-DR, IL1RAP, and TIM-3 (top). (B) Frequencies of LSC marker positive populations (CD123+ and TIM-3+) in bulk AML cells at diagnosis (middle) and relapse (right) compared with NBM cells (left) (NBM, n = 22-23; AML, n = 24-25). Mean ± standard error of the mean values are plotted. *P < .05; **P < .01; ***P < .001; ****P < .0001. NS, not significant.
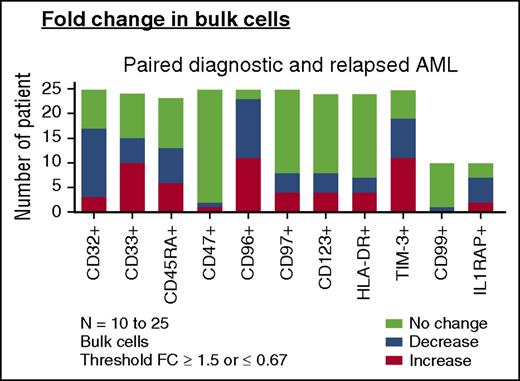
Change of LSC markers on paired AML diagnosis and relapse samples. Change in frequencies of LSC marker-positive bulk AML populations in paired diagnosis and relapse samples (AML, n = 23-25). For comparisons of change in frequency, a fold change (FC) cutoff of 1.5 was used in this analysis. “Increase” denotes any patient with FC ≥ 1.5 at relapse than at diagnosis. “Decrease” denotes any patient with FC of ≤ 0.67 at relapse than at diagnosis. “No change” denotes a FC <1.5 to >0.67 from diagnosis to relapse (bottom).
Change of LSC markers on paired AML diagnosis and relapse samples. Change in frequencies of LSC marker-positive bulk AML populations in paired diagnosis and relapse samples (AML, n = 23-25). For comparisons of change in frequency, a fold change (FC) cutoff of 1.5 was used in this analysis. “Increase” denotes any patient with FC ≥ 1.5 at relapse than at diagnosis. “Decrease” denotes any patient with FC of ≤ 0.67 at relapse than at diagnosis. “No change” denotes a FC <1.5 to >0.67 from diagnosis to relapse (bottom).
We then examined the expression of CD32, CD33, CD45RA, CD47, CD96, CD97, CD123, HLA-DR, and TIM-3 on functionally defined LSC and non-LSC populations (supplemental Figure 6). For the 12 non-LSC populations at diagnosis that developed LSC activity at relapse, only the frequency of TIM-3 and CD45RA increased (26.29% ± 24.66% vs 50.86% ± 33.37% [P = .0342] and 31.97% ± 25.75% vs 59.02% ± 34.41% [P = .0322], respectively). Thus, despite a 9- to 90-fold increase in LSC frequency at relapse as defined by LDA, the frequency of leukemia populations identified by previously reported LSC antigens either did not change or changed little from diagnosis to relapse.
Genomic changes after relapse
One potential mechanism for the increased LSC frequency and expanded phenotype (Table 1; Figure 1) at relapse would be the acquisition of additional cytogenetic events or mutations after therapy or the emergence of a refractory subclone. We analyzed pretherapy and posttherapy cytogenetic reports and performed targeted sequencing of 33 genes commonly mutated in hematologic malignancies on bulk cells (n = 18) or CD3−CD19− cells (n = 5) from 23 paired bone marrow or peripheral blood samples at diagnosis and in relapse (supplemental Table 1). Only disease-associated mutations were reported for this analysis. In 12 of the 22 patients for whom pretherapy and postrelapse cytogenetic studies were available was acquisition of additional cytogenetic abnormalities on relapse identified. Targeted sequencing identified 43 disease-associated mutations in 23 diagnostic samples and 47 disease-associated mutations in the paired relapse samples. Flt3 mutations represented the most common lost or acquired mutation (5 of 23 patients), followed by NRAS/KRAS. Of the 22 patients for whom both cytogenetic and targeted mutational analyses were available, 16 demonstrated the acquisition of additional genetic events at relapse. Four of the 7 patients for whom LSC functional analyses were performed demonstrated the acquisition of additional genetic events at relapse. Together, these data indicate significant genetic changes occur upon relapse of leukemic disease, suggesting that at least some of the changes in LSC frequency and diversity may derive from genetic instability and/or the selection of minor subclones.
To characterize the genomic heterogeneity across all LSC and non-LSC populations at diagnosis and after relapse, we performed targeted sequencing on sorted functionally defined LSC and non-LSC populations from paired diagnosis and relapse samples from 6 patients (supplemental Figure 7). For this analysis, we used the Illumina TruSight Myeloid Sequencing Panel (Illumina, San Diego, CA). This panel included a greater number of genes (64) in comparison with the bulk sample analyses (supplemental Table 1), and the libraries generated were sequenced to generate mean amplicon coverage depth of 15 000× or greater. All exonic calls compared with the reference standard were included for the analysis. For all patients except AML3, the majority of variants were detected in all sorted populations at diagnosis and relapse. For example, in AML15, only the CD34+CD38+ population at diagnosis demonstrated LSC activity, whereas at relapse, we demonstrated a 25-fold increase in LSC activity and all populations except the CD34−CD38− robustly engrafted recipient mice. Despite this large increase in LSC activity, the genetic profile of the 4 populations was generally stable between diagnosis to relapse arguing against genetic instability as the driver for the changes in the leukemic stem cell pool for this patient. For AML3, the 2 CD34 negative populations did not demonstrate LSC activity before treatment but acquired this activity at relapse. The genomic profile of these populations remained very stable between diagnosis and relapsed despite the large shift in LSC activity. These findings argue against large-scale genomic instability as the sole driver of the LSC expansion after treatment and relapse.
We then examined the genetic profiles of non-LSC populations before therapy that acquired LSC activity at relapse, as well as the LSC populations before treatment that retained LSC activity at relapse and identified those mutations either gained or lost. When comparing the sequencing data from paired diagnosis and relapse subpopulations, the most common variants that demonstrated a net gain at relapse involved DNMT3A, CDKN2A, HRAS, and PDGFRA. A DNMT3A variant not present or only present in 1 allele in a sorted population at diagnosis was detected or biallelic in 10 sorted populations from 4 patients at relapse. In 1 of the 2 patients that did not acquire a DNMT3A mutation by at least 1 sorted population at relapse, alternate DNMT3A variants were present and stable at diagnosis and relapse. Taken together, genomic analyses of both bulk and sorted populations (non-LSC and LSC) indicate a modest degree of genetic instability consistent with previous reports.27 However, no detectable genetic alteration explains the dramatic change in LSC frequency or acquisition of LSC potential by non-LSC populations.
High-dimensional mass cytometry identifies large-scale changes after chemotherapy and progression
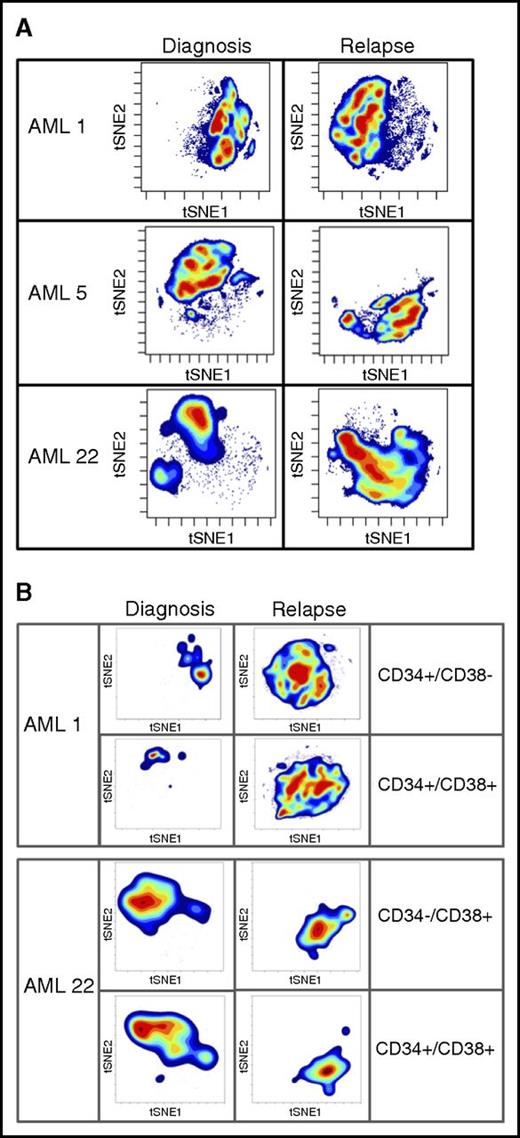
To further probe changes that may drive LSC expansion after relapse, we took advantage of high-dimensional mass cytometry (CyTOF). CyTOF allows for the simultaneous assessment of more than 60 surface and intracellular antigen profiles, including cell signaling. Coupling CyTOF with high-dimensional data analytics allows for the simultaneous assessment of multiple biological properties. We applied this approach to 2 cases in which functional LSC expansion was identified (Figure 4). This analysis revealed major differences between the diagnosis and relapse samples, as evidenced by the multiple changes in both cell surface and intracellular signaling pathways. To visualize these changes, we employed the viSNE algorithm, as shown in Figure 4. The viSNE plots are conceptually similar to principal component analyses and are used as a method of displaying high-dimensional single-cell data. Each position in the 2-dimensional plot represents proximity and similarity of single cells in a high-dimensional space.28 The maps in Figure 4 were generated according to cell surface phenotype, using 14 separate antigens. The Jensen-Shannon divergence test was used to compare the similarity of viSNE plots between diagnosis and relapse. The test confirmed striking differences, with scores of AML1- 0.67531, AML5- 0.6808, and AML22-0.67365. The Jensen-Shannon divergence score can range from 0 (no difference) to 1 (no similarity), with scores around 0.7 signifying major differences. The single antibody plots (Figures 2 and 3) reveal very few changes in frequency of expression of these markers. It is through high-dimensional analysis that global changes are recognized. The significant immunophenotype shifts are a result of acquisition or loss of specific markers when considered in the context of high-dimensional space. These changes are difficult to resolve using traditional flow, but mass cytometry brings such alterations to light. A more in-depth example of CyTOF-based analysis is shown in supplemental Figure 8. In this figure, by individually visualizing each antigenic marker in the context of a viSNE plot, we can detect population-based changes that occur at relapse. For this patient with AML, significant changes in the subpopulations that express p-p38, CD38, pNFkB, and CD56 are all readily evident. For example, using this type of high-dimensional analysis, one can easily see that the cells expressing CD38 at diagnosis are not the same as the cells expressing CD38 at relapse. Traditional flow cytometry would usually miss this type of subtle shift. However, when each change of this type is compiled into the cumulative data shown in Figure 4, substantial changes in phenotype are easily detectable. These data indicate that the large increase we observe in both LSC frequency and diversity is reflected in measurable changes in cellular physiology when a complex assay coupled with high-dimensional space data analytics are employed. Further analyses will be required to determine whether any particular phenotypic characteristic can be used to identify functionally defined LSCs.
High-dimensional mass cytometry of diagnosis and relapse specimens. (A) The viSNE maps of diagnosis and relapse specimens, as defined by cell surface antigens. High-dimensional mass cytometry was performed on 3 cases (AML1, AML5, and AML22) at diagnosis and relapse, where functional LSC expansion was identified by LDA. viSNE analysis was employed to display the single cell data results, using data from 14 separate cell surface antigens with a Jensen-Shannon Divergence score for pairs AML1, 0.67531; AML5, 0.6808; and AML22, 0.67365. These data indicate the large measurable changes in cellular physiology between diagnosis and relapse for the total mononuclear cell population. (B) The viSNE maps of functionally defined LSC populations at diagnosis and relapse; specimens as defined by cell surface antigens.
High-dimensional mass cytometry of diagnosis and relapse specimens. (A) The viSNE maps of diagnosis and relapse specimens, as defined by cell surface antigens. High-dimensional mass cytometry was performed on 3 cases (AML1, AML5, and AML22) at diagnosis and relapse, where functional LSC expansion was identified by LDA. viSNE analysis was employed to display the single cell data results, using data from 14 separate cell surface antigens with a Jensen-Shannon Divergence score for pairs AML1, 0.67531; AML5, 0.6808; and AML22, 0.67365. These data indicate the large measurable changes in cellular physiology between diagnosis and relapse for the total mononuclear cell population. (B) The viSNE maps of functionally defined LSC populations at diagnosis and relapse; specimens as defined by cell surface antigens.
Discussion
The cancer stem cell model is frequently cited to explain why the majority of patients relapse after initial therapy for AML. To date, there has been little effort to assess this supposition or to understand the effect of chemotherapy and relapse on the model. By employing limiting dilution analyses, the gold standard for assessing stem cell frequency, we were able to identify a previously unreported 9- to 90-fold increase in LSC activity after relapse. The increase in LSC frequency at relapse is associated with an expansion of LSC activity to populations of cells resembling more differentiated/mature progenitors, irrespective of the antigens used to define the populations. Importantly, the frequencies of populations defined by previously reported LSC antigens do not correlate with the postrelapse LSC expansion identified by LDA. High-dimensional flow cytometry reveals large-scale changes in leukemic blasts, as well as functionally defined LSC populations after relapse. Collectively, the data indicate that relapse after conventional chemotherapy is characterized by vastly more complex and heterogeneous LSC populations. We propose that this phenomenon lies at the root of clinical drug resistance and poor outcomes for patients who do not maintain remission after initial therapy. Indeed, both the increased frequency of LSC and their expanded diversity almost certainly contribute to much more drug-resistant/refractory disease.
In normal hematopoiesis, the capacity for self-renewal is retained within a rare relatively quiescent population of HSCs. As one moves further away from the HSC, CD34 and CD38 expression define the degree of differentiation and maturation with HSCs and early progenitors delineated as CD34+CD38− and late progenitors as CD34+CD38+.29 Studies in CML blast crisis and murine models for AML demonstrate that LSC population phenotypes are in part dependent on the stage of differentiation from which the fully transformed leukemia arises, as well as the oncogenic event or events; thus non-self-renewing normal hematopoietic elements may be transformed into self-renewing LSC populations.30 Adding to this complexity, recent reports from prechemotherapy and remission samples from patients with AML found that cells resembling HSCs harbor a subset of the mutations necessary for the development of AML.31-33 Our findings demonstrate that as the disease progresses (or responds to therapeutic insult), LSC activity increases and likely expands to additional compartments. Comparison of pretherapy and postrelapsed LSC surface antigen phenotypes demonstrates that CD34 and CD38, as well as most of the previously published markers for LSCs, are no longer capable of segregating LSC activity after relapse.
Our findings have implications for future therapeutics in cancer. There has been a considerable effort to identify CSC specific targets for developing therapeutics. In AML these include CD33, CD123, and CD47. The next generation of immuno-conjugates and CAR-T cells are now making their way to early clinical studies, using these antigens as targets. Although we did not directly assess each antigen’s ability to segregate functional LSC activity at diagnosis and relapse, our experience with CD32, CD34, and CD38 raises serious concerns about a therapeutic approach that relies on targeting a single antigen to eradicate CSCs, especially in the relapse setting, where most novel agents are initially employed. Furthermore, it is critical that the nature of LSCs at relapse be reflected in the design and evaluation of new therapeutic approaches. Ongoing studies will provide additional information as to the contributions of treatment and clonal selection on the dramatic changes we observe in the LSC pool after relapse. Current approaches to treating patients with AML, including those studied here, involve rapid administration of cytotoxic agents that are known to markedly stress malignant and normal hematopoiesis. However, with the advent of more advanced targeted drugs, it may be that therapies that avoid this stress to the LSC pool can yield comparable levels of disease control without inducing such dramatic changes in the nature of the LSC population. If so, this may leave patients with more options for secondary and tertiary lines of therapy extending disease control and enhancing their quality of life, especially for those patients whose age, pretreatment cytogenetics, and molecular profiles predict failure for standard approaches
In summary, our data support a central role for LSCs in the pathogenesis of AML. Given the dismal prognosis of relapsed AML, understanding how the LSC population evolves is critical to improving outcomes. Our examination of sorted populations using targeted sequencing suggest genomic instability is not the major driver of the observed expansion of LSC activity. Our initial studies applying high-dimensional mass cytometry to paired diagnostic and relapse samples reveals large-scale changes in the disease not detected using standard flow cytometry analysis. Additional global approaches such as the study of the epigenome and proteome, as well as single-cell analyses, will be required to identify the mechanisms by which this evolution occurs, as well as how these findings may apply to other malignancies.
Presented in abstract form at the 55th annual meeting of the American Society of Hematology, New Orleans, LA, December 10, 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Pam Iadarola for her administrative assistance; Tim Bushnell, Matthew Cochran, and the University of Rochester Medical Center Flow Core facility for maintenance of fluorescence-activated cell sorting facilities; Jessica Mavor and Laurie Ford for their assistance in obtaining patient samples at the J. P. Wilmot and the Roswell Park Cancer Institutes; and patients of at the J. P. Wilmot and the Roswell Park Cancer Institutes who donated tissue samples to this effort. This work is dedicated to the memory of M.W., whose contributions to this project were critical to its fruition.
This study was funded by National Institutes of Health National Cancer Institute 1R21CA149848-01, Wilmot Cancer Research Fellowship, Leukemia and Lymphoma Society Translational Grants (6230-11 and 6133-12), and the Empire State Institutional Training Program in Stem Cell Research.
Authorship
Contribution: T.-C.H., C.T.J., and M.W.B. designed the study; T.-C.H., M.L., and B.M.S. performed experiments; T.-C.H., M.L., B.M.S., J.M.A., J.R.M., C.T.J., and M.W.B. performed analysis; M.W. and E.S.W. contributed vital new reagents or analytical tools; T.-C.H., K.M.O., J.L.L., J.H.M., C.T.J., and M.W.B. wrote the manuscript; T.-C.H., B.M.S., J.M.A., K.M.O., J.L.L., J.H.M., M.G., J.D.M., J.Z., E.S.W., C.T.J., and M.W.B. revised the manuscript; and T.-C.H., M.L., B.M.S., J.M.A., J.R.M., K.M.O., J.L.L., J.H.M., M.G., J.D.M., J.Z., E.S.W., C.T.J., and M.W.B. approved the final version of the manuscript.
Conflict-of-interest disclosure: E.S.W. receives research funding from Immunogen. M.W.B. receives research funding from Millenium. J.M.A. owns stock of Pacific Biosciences. The remaining authors declare no competing financial interests.
Correspondence: Michael W. Becker, Wilmot Cancer Center, 601 Elmwood Ave, Box 704, Rochester, NY 14642; e-mail: michael_becker@urmc.rochester.edu.