To the editor:
Hypereosinophilic syndrome (HES) is a heterogeneous group of disorders characterized by (1) persistent peripheral eosinophilia, (2) target organ pathology mediated by infiltrating eosinophils, and (3) the absence of known infectious or allergic causes of hypereosinophilia.1,2 There are multiple variants, including primary, secondary, and idiopathic.1,2 The lymphocytic variant HES (L-HES) is a unique subtype of secondary HES characterized by eosinophilia and eosinophil-infiltrating lesions of the skin, soft tissue, and (rarely) internal organs.1,2 In L-HES, an abnormal T-cell clone (most commonly CD3−CD4+) produces pathological amounts of eosinophil-promoting T helper (Th)2 cytokines, such as interleukin (IL)-4, IL-5, and IL-13.1 These Th2 cytokines are thought to drive the polyclonal expansion of eosinophils in skin, subcutaneous tissue, and blood.1 To date, the molecular mechanisms underlying L-HES pathogenesis have been unclear. We hypothesized a genetic basis for this disorder. We therefore performed whole-exome sequencing of the T-cell clone and matched normal monocytes from a patient with L-HES. All human studies were approved by the institutional review boards at Yale School of Medicine and Johns Hopkins School of Medicine.
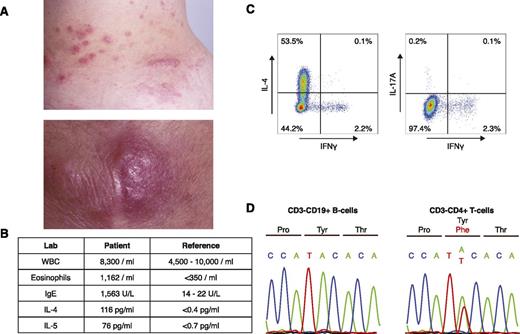
A 43-year-old woman initially presented with a 12-year history of persistent eosinophilia and eosinophil-infiltrating nodules of the skin, throat, and lymph nodes (Figure 1A-B). At diagnosis, laboratory testing revealed elevated serum IL-4 and IL-5, as well as a prominent CD3−CD4+ T-cell population (L-HES clone; 874/μL). Upon stimulation with phorbol 12-myristate 13-acetate (50 ng/mL) and ionomycin (1 μg/mL) for 5 hours, these cells produced IL-4 (a Th2 cytokine), but not interferon-γ (a Th1 cytokine) or IL-17 (a Th17 cytokine) (Th1/Th2/Th17 Phenotyping Kit; BD Pharmingen) (supplemental Table 1, available on the Blood Web site; Figure 1C). Clinical follow-up for the past 20 years revealed no progression to lymphoma or leukemia since the time of sample acquisition.
Identification of a p.Y640F STAT3 mutation in a patient with L-HES. (A) Clinical photographs of eosinophil-infiltrated papules and nodules on the patient’s trunk (upper) and elbow (lower). (B) Table of relevant laboratory findings, with reference values. (C) Intracellular cytokine staining of CD3−CD4+ T cells. Peripheral blood mononuclear cells were stimulated for 5 hours with phorbol 12-myristate 13-acetate (50 ng/mL) and ionomycin (1 μg/mL). Cells were subsequently fixed, permeabilized, and stained with a phycoerythrin/cyanin 7–labeled anti-human CD3 antibody (BD Pharmingen) and an antibody cocktail containing antibodies for CD4, IL-4, IL-17A, and interferon-γ (IFNγ) (BD Pharmingen). (D) Confirmation of the somatic STAT3 mutation in CD3−CD4+ T cells by Sanger sequencing. Matched normal CD3+CD19− B cells were used as a control.
Identification of a p.Y640F STAT3 mutation in a patient with L-HES. (A) Clinical photographs of eosinophil-infiltrated papules and nodules on the patient’s trunk (upper) and elbow (lower). (B) Table of relevant laboratory findings, with reference values. (C) Intracellular cytokine staining of CD3−CD4+ T cells. Peripheral blood mononuclear cells were stimulated for 5 hours with phorbol 12-myristate 13-acetate (50 ng/mL) and ionomycin (1 μg/mL). Cells were subsequently fixed, permeabilized, and stained with a phycoerythrin/cyanin 7–labeled anti-human CD3 antibody (BD Pharmingen) and an antibody cocktail containing antibodies for CD4, IL-4, IL-17A, and interferon-γ (IFNγ) (BD Pharmingen). (D) Confirmation of the somatic STAT3 mutation in CD3−CD4+ T cells by Sanger sequencing. Matched normal CD3+CD19− B cells were used as a control.
We isolated the patient’s aberrant T-cell clone (CD3−CD4+ T cells) and matched normal monocytes (CD3−CD14+ cells) using flow cytometry (supplemental Table 1). Exome sequencing and subsequent identification of somatic mutations were performed as previously described.3 We identified 1 focal region of acquired copy-neutral loss of heterozygosity (chromosome 6: 64,752,218-114,485,941) and 25 somatic single-nucleotide mutations. We failed to identify any copy-number losses or gains.
We identified a somatic gain-of-function mutation in STAT3 (p.Y640F, c.1919A>T) in the CD3−CD4+ T cell that was not present in the matched normal CD3−CD14+ monocytes. This mutation was confirmed using Sanger sequencing (Figure 1D). Previously identified in the CD8+ lymphoproliferative disease T-cell large granular leukemia,4,5 the p.Y640F mutation alters a conserved tyrosine residue in the Src homology 2 domain of signal transducer and activator of transcription (STAT)3. STAT3 (p.Y640F) is a functionally validated gain-of-function mutation that has been shown to render STAT3 constitutively active in multiple cell lines (kidney epithelial cells,5 hepatic epithelial cells,6 fibroblasts,7 prostate cancer cells,7 and lung cancer cells7 ). There were no somatic mutations in other consensus cancer genes8 or genes underlying inherited immune disorders.9
To assess the generalizability of our findings, we expanded our cohort to include 2 additional patients with L-HES. Similar to our index case, these patients had erythematous, edematous, and pruritic cutaneous and subcutaneous papules and nodules with a phenotypically abnormal CD3−CD4+ T-cell clone. The prevalences of CD3−CD4+ T cells (L-HES clones) in these 2 patients were 1% and 5%.
Targeted DNA sequencing of STAT3 in L-HES clones from these patients did not reveal evidence of known activating STAT3 mutations (supplemental Table 2; data not shown). Nevertheless, we hypothesized that STAT3 activation was a common feature to all 3 cases of L-HES. To test this, we first generated an unbiased STAT3 gene signature from chromatin immunoprecipitation sequencing data sets10-12 (supplemental Table 3). This signature consisted of genes that harbored STAT3 binding sites in 4 data sets: CD4+ T lymphocytes,10,11 ES cells,12 and breast cancer cells (unpublished data). Genes that were not expressed in CD4+ T cells (fragments per kilobase per million reads <0.1) were excluded from this gene signature.
Using standard library preparation and sequencing protocols,3 we then performed RNA sequencing of the 3 L-HES clones and memory CD4+ T cells from 3 unrelated healthy individuals. We found significant enrichment of the STAT3 signature genes in each of the 3 L-HES samples. The transcript levels of STAT3 signature genes were significantly upregulated in L-HES clones compared with CD4+ T cell controls (mean fold-change values were 2.0, 3.3, and 3.7). Using a previously described algorithm,3 we found that this enrichment of STAT3 gene targets was unlikely to occur by chance alone (P < .008, binomial distribution; Figure 2A). We confirmed these findings with an independent algorithm (gene set enrichment analysis; http://www.broadinstitute.org/gsea/index.jsp)13 (P = .002; Figure 2B).
Activation of STAT3 gene targets in L-HES clones. (A) The mean fold-change of transcript levels (log2) in L-HES clones compared with CD4+ T-cell controls (control memory [CM]) as evaluated through RNA sequencing. L-HES clone 1 possesses the p.Y640F mutation. ***P = .0002; **P = .0007; *P = .008. (B) Gene set enrichment analysis of STAT3 gene signature in L-HES clones compared with CD4+ T-cell controls (P = .002). (C) Relative levels of messenger RNA (mRNA) transcripts of known STAT3 gene targets in unstimulated L-HES clones as compared with CD4+ T-cell controls. (D) Relative levels of cytokine expression in L-HES clones as compared with CD4+ T-cell controls. For panels C-D, closed symbols represent the L-HES clone containing the p.Y640F mutation. In panel D cytokine profiles, unstimulated L-HES clones were not annotated with closed symbols for the sake of clarity. Each T-cell sample was cultured with or without pharmacologic stimulation with phorbol 12-myristate 13-acetate and ionomycin. *P < .05, **P < .005, ***P < .0005. fpkm, fragments per kilobase per million reads.
Activation of STAT3 gene targets in L-HES clones. (A) The mean fold-change of transcript levels (log2) in L-HES clones compared with CD4+ T-cell controls (control memory [CM]) as evaluated through RNA sequencing. L-HES clone 1 possesses the p.Y640F mutation. ***P = .0002; **P = .0007; *P = .008. (B) Gene set enrichment analysis of STAT3 gene signature in L-HES clones compared with CD4+ T-cell controls (P = .002). (C) Relative levels of messenger RNA (mRNA) transcripts of known STAT3 gene targets in unstimulated L-HES clones as compared with CD4+ T-cell controls. (D) Relative levels of cytokine expression in L-HES clones as compared with CD4+ T-cell controls. For panels C-D, closed symbols represent the L-HES clone containing the p.Y640F mutation. In panel D cytokine profiles, unstimulated L-HES clones were not annotated with closed symbols for the sake of clarity. Each T-cell sample was cultured with or without pharmacologic stimulation with phorbol 12-myristate 13-acetate and ionomycin. *P < .05, **P < .005, ***P < .0005. fpkm, fragments per kilobase per million reads.
We then searched for STAT3-dependent candidate genes that contribute to the unique immunophenotype of L-HES. Previous work suggested that STAT3 activation is both necessary and sufficient for Th2 differentiation.14 The investigators found that STAT3 upregulates the expression of transcription factors critical for Th2 differentiation (ie, GATA-3, MAF, and BATF).14 In L-HES clones, we found significant upregulation of GATA-3 (12.2-fold; P = .002, unpaired Student t test) and BATF (7.4-fold; P = .006, unpaired Student t test) (supplemental Table 4), which are both required for Th2 differentiation. MAF was not upregulated (Figure 2C). Strikingly, overexpression of GATA-3 alone has been shown to be sufficient to induce Th2 differentiation and subsequent expression of Th2 cytokines.15
Lastly, we analyzed RNA sequencing to elucidate cytokine profiles of L-HES clones. For these experiments, L-HES clones were either unstimulated or stimulated with phorbol 12-myristate 13-acetate (50 ng/mL) and ionomycin (1 μg/mL) for 5 hours. Unstimulated and stimulated memory CD4+ T cells from 3 healthy individuals served as controls. As previously described,14 we found that stimulated CD3−CD4+ T cells from L-HES patients expressed high levels of Th2 cytokines. IL-4 and IL-13 were both >130-fold higher than stimulated control memory CD4+ T cells (P < .005; unpaired Student t test). IL-5 was 50-fold higher in L-HES clones, but this difference failed to reach statistical significance (P = .06). L-HES clones also produced significantly lower amounts of interferon-γ and IL-17 (Figure 2D; supplemental Table 5). Unstimulated L-HES cells exhibited a similar, albeit more modest, upregulation of Th2 cytokines compared to unstimulated control memory CD4+ T cells (supplemental Table 6).
Our data suggest that disease-relevant STAT3 gene targets are overexpressed in L-HES clones. A functionally validated, gain-of-function mutation in STAT3 activated the pathway in 1 patient in our cohort. STAT3 activation in other patients may be a result of genetic mutations or epigenetic changes in other components of the Janus kinase (JAK)-STAT pathway. Importantly, these results collectively suggest that JAK-STAT inhibitors, such as the JAK inhibitors and STAT3 inhibitors currently in clinical trials,16 may be an effective treatment of L-HES.
Lastly, this discovery expands the spectrum of immunologic diseases associated with activating STAT3 mutations. The phenotypes associated with STAT3 mutations appear to depend on the cell type (or types) harboring the mutations. When present in CD8+ T cells, these mutations promote T-cell large granular leukemia.4,5 When present in the germ line (ie, in all immune compartments), activating STAT3 mutations cause early-onset multiorgan autoimmune syndromes.17 We propose that activating STAT3 mutations, when present in circulating memory CD4+ T cells, lead to L-HES.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The work was supported by the Howard Hughes Medical Institute (R.P.L.), the Yale Specialized Programs of Research Excellence Career Development Award (J.C.), an American Cancer Society pilot grant (IRG-58-012-57) (J.C., T.W.), National Institutes of Health National Cancer Institute grant 1K08CA191019 (J.C.), and the Agency for Science, Technology, and Research, Singapore (G.G.).
Contribution: J.C. and R.P.L. designed the study, analyzed the data, and wrote the manuscript; J.C., J.R.T., S.W., C.W., T.W., J.L.L., and G.G. performed the bioinformatics analysis; J.C., C.W., and B.S.H. performed the flow cytometry experiments; and J.C., C.W., A.S., S.R.L., W.R.H., E.C.V., and B.A.K. prepared the clinical summary.
Conflict-of-interest disclosure: B.A.K. has served on an advisory board for Pfizer. The remaining authors declare no competing financial interests.
Correspondence: Jaehyuk Choi, Department of Dermatology, Northwestern University Feinberg School of Medicine, Robert H. Lurie Medical Research Center 5-115, 303 E. Superior St, Chicago, IL 60611; e-mail: jaehyuk.choi@northwestern.edu.

![Figure 2. Activation of STAT3 gene targets in L-HES clones. (A) The mean fold-change of transcript levels (log2) in L-HES clones compared with CD4+ T-cell controls (control memory [CM]) as evaluated through RNA sequencing. L-HES clone 1 possesses the p.Y640F mutation. ***P = .0002; **P = .0007; *P = .008. (B) Gene set enrichment analysis of STAT3 gene signature in L-HES clones compared with CD4+ T-cell controls (P = .002). (C) Relative levels of messenger RNA (mRNA) transcripts of known STAT3 gene targets in unstimulated L-HES clones as compared with CD4+ T-cell controls. (D) Relative levels of cytokine expression in L-HES clones as compared with CD4+ T-cell controls. For panels C-D, closed symbols represent the L-HES clone containing the p.Y640F mutation. In panel D cytokine profiles, unstimulated L-HES clones were not annotated with closed symbols for the sake of clarity. Each T-cell sample was cultured with or without pharmacologic stimulation with phorbol 12-myristate 13-acetate and ionomycin. *P < .05, **P < .005, ***P < .0005. fpkm, fragments per kilobase per million reads.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/7/10.1182_blood-2015-06-654277/4/m_948f2.jpeg?Expires=1769081909&Signature=RidL3WCgx43DFwgrn89RtfwtgQrv6~PqpnlT1kikup4NDmrNIk0ZFujQQPnJVQfC3tfWtunvRFbFePOIQqO3TGTPMCfLt0cfnqOLPYLkr7iIntNlfQu390f3cNJjkmN8oEmWf6KbFYX91VuzPU6x8XIWpEmzoXqGZj06dbobbTp9ct1eZqnk-xCDSC5WkMBOFdUFf710GwFU49l-ZTbzFuR-350-gx9EbHsi~SS~FWGSYaqPVLeGwtDK-s4sGfOKI14o4owZYzfpRx~tDzkl1ZZxavBiuyw-KoPDUZTMlSHD8ccgsNbelfleBYwInQx7L~1gS19NUPGXoPG-c8pwJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal