Key Points
Mef2c and Mef2d are activated by the pre-B-cell receptor and are essential for pre-B-cell transition.
Mef2c complexes with B-cell transcription factors to shut down the immediate early response and to initiate a new transcriptional network.
Abstract
The sequential activation of distinct developmental gene networks governs the ultimate identity of a cell, but the mechanisms involved in initiating downstream programs are incompletely understood. The pre-B-cell receptor (pre-BCR) is an important checkpoint of B-cell development and is essential for a pre-B cell to traverse into an immature B cell. Here, we show that activation of myocyte enhancer factor 2 (Mef2) transcription factors (TFs) by the pre-BCR is necessary for initiating the subsequent genetic network. We demonstrate that B-cell development is blocked at the pre-B-cell stage in mice deficient for Mef2c and Mef2d TFs and that pre-BCR signaling enhances the transcriptional activity of Mef2c/d through phosphorylation by the Erk5 mitogen-activating kinase. This activation is instrumental in inducing Krüppel-like factor 2 and several immediate early genes of the AP1 and Egr family. Finally, we show that Mef2 proteins cooperate with the products of their target genes (Irf4 and Egr2) to induce secondary waves of transcriptional regulation. Our findings uncover a novel role for Mef2c/d in coordinating the transcriptional network that promotes early B-cell development.
Introduction
B-cell development can be divided into a series of developmental stages that are characterized by the activation and repression of sequential transcriptional programs. To achieve this, differentiation steps are tightly regulated by intercellular signals and specific transcription factor (TF) networks, the interplay of which is poorly understood. Generation of the earliest B-cell progenitors depends on signaling mediated by the Flt3 ligand and interleukin 7 (IL7) and on TFs (E2a, Foxo1, Ikaros, and Runx1) that prime the B-cell program and (in cooperation with B-cell–specific factors [Ebf1 and Pax5]) activate the B-cell gene expression program and repress lineage-inappropriate genes.1-5 V(D)J gene rearrangements are initiated by Rag1/2 proteins during this early pro-B-cell stage, leading to the generation of immunoglobulin (Ig) heavy chains (IgH). The pre-B-cell stage marks the first important checkpoint of B-cell development. Association of newly generated IgH with the surrogate light chain (SLC) results in pre-B-cell receptor (pre-BCR) assembly and signaling via phosphatidylinositol-3-OH kinase (PI3K) and mitogen-activated protein kinases (MAPKs), triggering clonal expansion of pre-B cells expressing a functional pre-BCR.6-9
In addition to its mitogenic stimulus, pre-BCR signaling initiates a series of events that promote differentiation, including exit from cell cycle, downregulation of SLC gene expression, and rearrangements of Ig light chain (IgL) genes. The transcriptional network that initiates this process is only sketchily understood. The pre-BCR signaling induces expression of the Irf4 TF by an uncharacterized mechanism, which in turn stimulates the expression of Ikzf1 and Ikzf3 (encoding Ikaros and Aiolos, respectively), important suppressors of SLC expression.10,11 E2a and Foxo proteins are also known to induce IgL gene rearrangements through the activation of Rag1/2 expression, a critical step for the transition from the small pre-B cell to the immature B-cell stage.7,12 Replacement of SLC with IgL leads to BCR expression and marks the beginning of distinct genetic programs that drive the disparate functions of mature B cells.13-15
Mef2c, a member of the myocyte enhancer factor 2 (Mef2) family of TFs, has been previously implicated as a regulator of B-cell homeostasis and antigen activation,16-18 but its role in early B-cell development is unknown. The Mef2 proteins belong to the MADS-box family, which includes 4 paralogues in vertebrates (Mef2a-d) with a highly conserved DNA-binding domain.19 These factors are essential for muscle and neuronal development and are highly responsive to extracellular signaling pathways.20 Mef2c and Mef2d are highly expressed throughout B-cell differentiation.21,22 Furthermore, deletions of MEF2C and chromosomal translocations involving the MEF2D gene have been reported in acute lymphoblastic leukemia.23-25 We thus sought to determine whether Mef2c and/or Mef2d play a role in early B-cell development. Utilizing knockout (KO) mice, in vitro cell systems, and genome-wide transcriptome and TF occupancy data, we demonstrate that Mef2c and Mef2d exhibit essential functions during pre-B-cell development through integrating pre-BCR signaling with transcriptional networks that promote transition to the immature stage.
Methods
Experimental animals and cell lines
The B6.129-Mef2cfl/fl, B6.129-Mef2d−/−, and B6.129-Cd79ahCre/+ (mb1-Cre) mouse strains26-28 were crossed to generate control mice and double knockouts, in which Mef2d is constitutively inactivated and Mef2c inactivation occurs in B-cell specified common lymphoid progenitors (Ly6d+).5 Cre-mediated excision was confirmed by polymerase chain reaction (PCR) (supplemental Figure 1, available on the Blood Web site). The Klf2Δ/ΔCd79ahCre/+ mice have been previously described.29 Mice were maintained at the Heinrich-Pette-Institute Animal Facility; the Hamburg Commission for Animal Experiments approved all studies.
Murine BMiFLT3(15-3) pro-B cells5 were cultivated in modified Eagle medium (MEM; Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS). Blnk-deficient pre-B cells30 were cultivated in Iscove modified Dulbecco medium (IMDM; Life Technologies) supplemented with 10% FCS, 50 µM β-mercaptoethanol, and 5 ng/mL IL7. The tamoxifen-inducible Blnk-ERt2 retroviral construct has been described previously.31
Flow cytometry analysis
Single-cell suspensions for fluorescence-activated cell sorter (FACS) analysis and sorting were prepared from indicated organs from 7- to 14-week-old mice. Cells were stained with fluorophore- or biotin-conjugated antibodies (supplemental Table 1) and analyzed or sorted by flow cytometry (Canto-I/-II or Aria-I; BD Biosciences). A lineage cocktail (Lin−) was used to eliminate non-B cells. FACS data were analyzed with FACSDiva software (BD Biosciences).
Immunoblotting and antibodies
Cells were incubated for 1 hour with mitogen-activated protein kinase kinase (MEK) inhibitor U0126 (Cell Signaling) before harvesting. Preparation of lysates, immunoprecipitation, and immunoblotting was performed as previously described.32 Proteins were quantified using the Odyssey Image scanner system (LI-COR Biosciences) on membranes incubated with primary antibody overnight and then probed with IRdye 700– or 800–conjugated secondary antibodies. A list of antibodies is provided in supplemental Table 2.
Histology
Lymph nodes were fixed using formalin, embedded in paraffin, sectioned (4 μm), and stained with unconjugated rat anti-murine B220/CD45R antibody (clone RA3-6B2; BD Pharmingen) (1:400) followed by a peroxidase-conjugated second-step antibody using standard immunohistochemical procedures. Peroxidase activity was visualized with 3,3′-diaminobenzidine and sections were counterstained with hematoxylin. Whole lymph node sections were captured on a Zeiss microscope (Axioplan2 imaging) with ProgRes Capture Pro V2.8.8 imaging software.
RNA analysis
For quantitative reverse transcription PCR (RT-PCR) analysis, RNA was isolated using the PeqGOLD TriFast method (PeqLab Biotechnology) and analyzed using the Roche480 LightCycler with SYBR Green I master (Roche Diagnostics) and primer pairs presented in supplemental Table 3. For RNA-seq analysis, messenger RNA (mRNA) was isolated using the NEBNext Poly(A) mRNA Magnetic Isolation NEB module; libraries were generated using the TruSeq: RNA Sample Preparation V2 Low Throughput kit (Illumina) or the NEBNext Ultra RNA Library Prep kit for Illumina (both from New England Biolabs). Sequencing was performed on a HiSequation 2500 system (Illumina). Transcriptome analysis of pro-B cells obtained from Mef2c/dΔ/ΔCd79ahCre/+ (n = 3 per sort) or Cd79ahCre/+ control mice (n = 2 per sort) was performed on 3 biological replicas. For Klf2 functional analysis, transcriptomes were analyzed from either pro-B cells isolated from Klf2Δ/ΔCd79ahCre/+ (n = 2) or Cd79ahCre/+ (n = 2) littermates or from established pro-B cells retrovirally transduced to express Klf2. Reads (35-90 million per sample) were mapped to the mouse genome (mm10) using CLC Genomics Workbench v6.5.1 software (Qiagen). Analysis of differential gene expression was performed with the Baggerley statistical test procedure of the CLC Genomics Workbench software with all parameters at their default settings. RNA-seq data are available at the NCBI Gene Expression Omnibus (GEO; GSE73563).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) was performed twice on BMiFLT3(15-3) pro-B cells (1 × 107 cells per ChIP), as previously described.5 In brief, extracted DNA fragments were isolated, sequenced on a HiSequation 2500 (Illumina), and mapped to the mm9 assembly. Details for ChIP-Seq, motif, and distance analysis and associated references are available in supplemental Methods. ChIP-Seq data are available at GEO (GSE73563).
Reporter gene assay
A 1092-bp fragment of the Klf2 promoter was cloned into the pGL4.10 vector (Promega) containing the firefly luciferase gene. Cos7 cells were plated at a density of 4 × 104 per well in 24-well plates 1 day before DNA transfection (5 µg) by the calcium-phosphate method. Reporter gene assays were performed 48 hours after transfection using the Dual-Luciferase Reporter Assay Kit (Promega) and normalized using cotransfected renilla luciferase. Transfections were performed in duplicate in at least 2 independent experiments.
Results
Impaired large to small pre-B-cell transition after loss of Mef2c and Mef2d
To investigate the influence of Mef2c and Mef2d on B-cell development, we analyzed mice deficient for either Mef2d (Mef2dΔ/Δ) or Mef2c (Mef2cΔ/Δ), or for both (Mef2c/dΔ/Δ). B-cell development in the bone marrow (BM) of Mef2c or Mef2d single KO mice was similar to that of control littermates (Mef2c/d+/+) (Figure 1A). In contrast, significantly reduced numbers of small pre-B cells and cells of subsequent developmental stages were observed in double KOs (Figure 1A), although the pro-B- and large pre-B-cell compartments were only moderately affected in cell numbers (Figure 1A) and showed normal levels of IgH rearrangement (supplemental Figure 2). Consistent with those findings, CD19-positive mature B cells were barely detectable in the blood and spleen of Mef2c/d-deficient mice and histology staining of lymph nodes confirmed the absence of B-cell zones and low levels of B cells (Figure 1B-D). The observed block at the transition through the pre-B stage suggests that both Mef2c and Mef2d are required for progression of B-lymphoid precursors through the pre-BCR checkpoint (Figure 1E).
Functional deletion of Mef2c/d in B-cell progenitors leads to a developmental block of B-cell differentiation. (A) Flow cytometric analysis to detect and enumerate pro-B cells (Lin−CD19+B220medCD43hiIgL−), pre-B cells (Lin−CD19+B220medCD43med/loIgL−), immature B cells (Lin−CD19+B220medCD43med/loIgL+), and mature (recirculating) B cells (Lin−CD19+B220hiCD43−IgL+) in the BM of Mef2cΔ/Δ (light blue), Mef2dΔ/Δ (green), Mef2c/dΔ/Δ (red), and control mice (Mef2c/d+/+, dark blue). Pre-B cells were separated by forward light scatter (FSC) into large and small pre-B cells, as indicated. Representative flow cytometry plots of Mef2c/dΔ/Δ and Mef2c/d+/+ mice are shown (top). Graph (bottom) depicts the number of different B-cell subtypes within total BM. Each mouse is represented by a dot (n ≥ 8 per genotype). Horizontal lines indicate the median. **P < .01; ***P < .001; ns, not significant. (B-C) Evaluation of B cells in blood (n = 5 per cohort) (B) and spleen (n = 12 per cohort) (C) of Mef2c/dΔ/Δ and control mice (Mef2c/d+/+) using FACS analysis. Graph depicts the percentage of CD19+B220+ B cells. Dots represent individual mice and bars indicate the median. (D) B220/CD45R staining of B-cell zones (indicated by a star [★]) from lymph nodes of Mef2c/dΔ/Δ and Mef2c/d+/+ mice. B-cell zones are lacking in lymph nodes of Mef2c/dΔ/Δ mice. Representative stainings from 2 experiments (I and II) using 2 mice per group are shown. Bar, 200 µm. (E) Schematic representation of the observed B-cell differentiation block at the pre-B-cell stage in BM of Mef2c/dΔ/Δ mice. The stage in B-cell development at which Cd79a-hCre–mediated excision occurs is indicated. Receptors that are important for the proliferative expansion of the specific B-cell types are depicted.
Functional deletion of Mef2c/d in B-cell progenitors leads to a developmental block of B-cell differentiation. (A) Flow cytometric analysis to detect and enumerate pro-B cells (Lin−CD19+B220medCD43hiIgL−), pre-B cells (Lin−CD19+B220medCD43med/loIgL−), immature B cells (Lin−CD19+B220medCD43med/loIgL+), and mature (recirculating) B cells (Lin−CD19+B220hiCD43−IgL+) in the BM of Mef2cΔ/Δ (light blue), Mef2dΔ/Δ (green), Mef2c/dΔ/Δ (red), and control mice (Mef2c/d+/+, dark blue). Pre-B cells were separated by forward light scatter (FSC) into large and small pre-B cells, as indicated. Representative flow cytometry plots of Mef2c/dΔ/Δ and Mef2c/d+/+ mice are shown (top). Graph (bottom) depicts the number of different B-cell subtypes within total BM. Each mouse is represented by a dot (n ≥ 8 per genotype). Horizontal lines indicate the median. **P < .01; ***P < .001; ns, not significant. (B-C) Evaluation of B cells in blood (n = 5 per cohort) (B) and spleen (n = 12 per cohort) (C) of Mef2c/dΔ/Δ and control mice (Mef2c/d+/+) using FACS analysis. Graph depicts the percentage of CD19+B220+ B cells. Dots represent individual mice and bars indicate the median. (D) B220/CD45R staining of B-cell zones (indicated by a star [★]) from lymph nodes of Mef2c/dΔ/Δ and Mef2c/d+/+ mice. B-cell zones are lacking in lymph nodes of Mef2c/dΔ/Δ mice. Representative stainings from 2 experiments (I and II) using 2 mice per group are shown. Bar, 200 µm. (E) Schematic representation of the observed B-cell differentiation block at the pre-B-cell stage in BM of Mef2c/dΔ/Δ mice. The stage in B-cell development at which Cd79a-hCre–mediated excision occurs is indicated. Receptors that are important for the proliferative expansion of the specific B-cell types are depicted.
Identification of important Mef2c/d target genes at the pro-B-/pre-B-cell stage
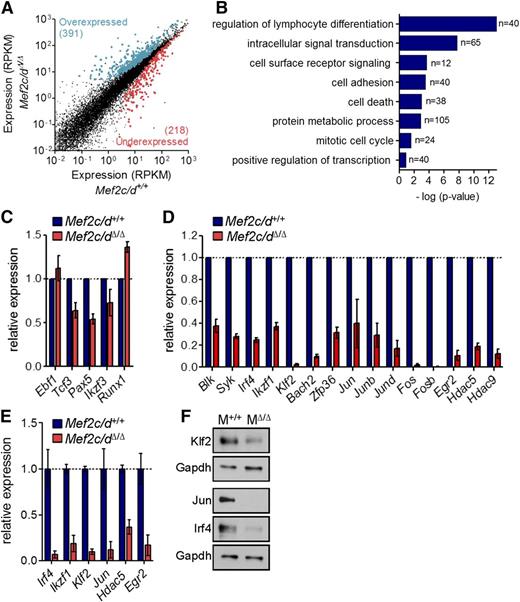
To identify Mef2c/d-regulated genes in early B-cell development, genome-wide gene expression profiles of wild-type and Mef2c/d-deficient pro-B cells (Lin−CD19+CD43highB220low) were analyzed by RNA-seq analysis. In comparison with controls, we found 609 genes significantly deregulated (≥1.5-fold; false discovery rate corrected P value < .05) in Mef2c/dΔ/Δ B-cell progenitors in 3 independent experiments (Figure 2A; supplemental Table 4). Gene ontology analysis using the STRING database33 was used to determine potential biological functions that are enriched in the differentially expressed genes (Figure 2B). Consistent with a critical role in B-cell development, the highest enrichment was seen in genes associated with lymphocyte differentiation. Strikingly, 65 of the deregulated genes belong to intracellular signaling pathways and 40 with cell adhesion. Gene products regulating metabolic processes and the mitotic cell cycle were also highly enriched (Figure 2B).
Genome-wide identification of Mef2 target genes using RNA-seq. (A) Scatter plot of differential gene expression (>1.5-fold; false discovery rate corrected P ≤ .05) in pro-B cells of Mef2c/dΔ/Δ vs control mice (Mef2c/d+/+) in 3 independent experiments. Lin−CD19+CD43highB220low pro-B cells were sorted by flow cytometry for preparation of RNA followed by deep sequencing. Gene expression is presented as normalized expression data (reads per kilobase of transcript per million reads mapped). Each dot represents 1 gene. Colors indicate overexpressed (blue) and underexpressed (red) genes in Mef2c/dΔ/Δ mice in relation to control mice. (B) Functional classification of genes significantly deregulated in pro-B cells of Mef2c/dΔ/Δ mice. Gene ontology (GO) analysis using the STRING database was performed to identify biological processes that are enriched in deregulated genes. The number of genes per class and the statistical significance is indicated. (C-D) Relative transcript levels determined by RNA-seq of genes encoding B-cell TFs (C) and a selection of differentially regulated genes (D) in pro-B cells of Mef2c/dΔ/Δ mice vs Mef2c/d+/+ control mice are depicted. Shown is the mean value of 3 independent experiments relative to controls. Error bars denote ± standard deviation (SD). (E) Quantitative RT-PCR (qRT-PCR) validation of transcript levels of a number of deregulated genes in Mef2c/dΔ/Δ mice vs Mef2c/d+/+ mice. qRT-PCR data were normalized to Hprt expression. Error bars represent ± SD. Data were combined from at least 2 independent experiments. (F) Immunoblot analysis of whole-cell lysates of Mef2c/d-deficient (MΔ/Δ) primary pro-B/pre-B cells (Lin−CD19+Igκ/λ−) in relation to pro-B/pre-B cells of Mef2c/d+/+ mice (M+/+). Protein levels of Klf2, Jun, and Irf4 are shown. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) served as a control. Data are representative of 2 independent experiments.
Genome-wide identification of Mef2 target genes using RNA-seq. (A) Scatter plot of differential gene expression (>1.5-fold; false discovery rate corrected P ≤ .05) in pro-B cells of Mef2c/dΔ/Δ vs control mice (Mef2c/d+/+) in 3 independent experiments. Lin−CD19+CD43highB220low pro-B cells were sorted by flow cytometry for preparation of RNA followed by deep sequencing. Gene expression is presented as normalized expression data (reads per kilobase of transcript per million reads mapped). Each dot represents 1 gene. Colors indicate overexpressed (blue) and underexpressed (red) genes in Mef2c/dΔ/Δ mice in relation to control mice. (B) Functional classification of genes significantly deregulated in pro-B cells of Mef2c/dΔ/Δ mice. Gene ontology (GO) analysis using the STRING database was performed to identify biological processes that are enriched in deregulated genes. The number of genes per class and the statistical significance is indicated. (C-D) Relative transcript levels determined by RNA-seq of genes encoding B-cell TFs (C) and a selection of differentially regulated genes (D) in pro-B cells of Mef2c/dΔ/Δ mice vs Mef2c/d+/+ control mice are depicted. Shown is the mean value of 3 independent experiments relative to controls. Error bars denote ± standard deviation (SD). (E) Quantitative RT-PCR (qRT-PCR) validation of transcript levels of a number of deregulated genes in Mef2c/dΔ/Δ mice vs Mef2c/d+/+ mice. qRT-PCR data were normalized to Hprt expression. Error bars represent ± SD. Data were combined from at least 2 independent experiments. (F) Immunoblot analysis of whole-cell lysates of Mef2c/d-deficient (MΔ/Δ) primary pro-B/pre-B cells (Lin−CD19+Igκ/λ−) in relation to pro-B/pre-B cells of Mef2c/d+/+ mice (M+/+). Protein levels of Klf2, Jun, and Irf4 are shown. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) served as a control. Data are representative of 2 independent experiments.
The gene expression pattern of a number of known B-cell TFs (Ebf1, E2a, Pax5, Aiolos) was only moderately affected or not changed (Figure 2C). However, transcript levels of several genes encoding modulators of the pre-B-cell signaling pathway or downstream effectors were significantly reduced by a factor >2 in Mef2c/d-deficient pro-B cells, including tyrosine kinases (Blk, Syk)7,34 and the TFs Irf4 (Irf4),10 Ikaros (Ikzf1),10,11 Bach2 (Bach2),35 and Krüppel-like factor 2 (Klf2)34 (Figure 2D; supplemental Table 4). A striking reduction in expression was observed in several immediate early genes (IEGs) (Figure 2D). Predominant were not only members of the AP1 family (Jun/Junb/Jund, Fos/Fosb), but also the early growth response factor Egr2 and the zinc finger protein 36 (Zfp36). Furthermore, significantly reduced transcript levels of genes encoding the class II histone deacetylase family (Hdac5/9) were detected. Transcript levels for a selection of these genes were confirmed using quantitative RT-PCR (Figure 2E). Significantly reduced protein levels of Klf2, Jun, and Irf4 were also detected in Mef2c/d KO pro-B cells (Figure 2F).
Genome-wide identification of Mef2 direct target genes
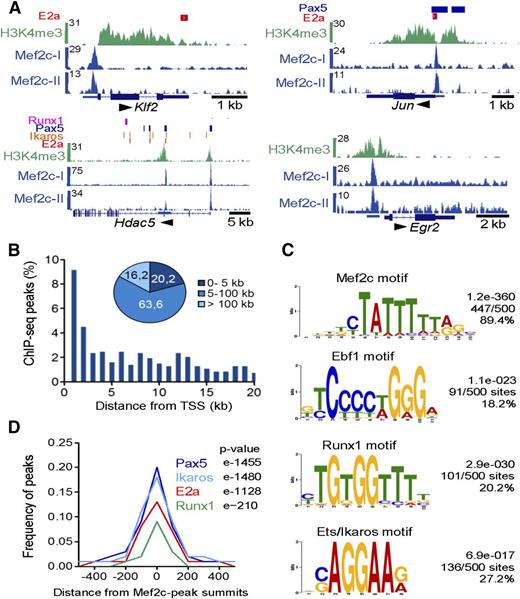
To determine which of the differentially expressed genes were indeed direct Mef2 targets, we examined the genome-wide binding pattern of Mef2c in transformed murine pro-B cells using ChIP-seq. We identified 725 binding sites of Mef2c (supplemental Table 5). Mef2c-occupied sites were detected at regulatory regions of genes encoding hematopoietic transcriptional regulators Klf2 and Runx1, the histone deacetylase family II (Hdac5/9), as well as genes encoding the IEG family members Jun, Fos, Fosb, Erg2, and Kfp36 (Figure 3A; supplemental Figure 3A). A second ChIP-seq analysis confirmed Mef2c-binding sites for these genes (Figure 3A; supplemental Figure 3A; supplemental Table 5). Interestingly, 9% of Mef2c-binding sites mapped within 1 kb of the transcriptional start site (TSS) of annotated genes and nearly 85% within 100 kb of the TSS, indicating that Mef2c binds to both promoter and enhancer regions of target genes in pro-B cells (Figure 3B). An example of long-distance enhancer regions bound by Mef2c was found flanking the Irf4 gene and its neighbor Dusp22, both differentially deregulated in the double KOs and upregulated during the pre-B-cell transition (supplemental Figure 3A).
Genome-wide mapping of direct target genes of Mef2c identified by ChIP-seq analysis in pro-B cells. (A) Mef2c bound regions in 4 gene loci identified by peak calling. Known binding sites of E2a, Runx1, and regions with H3K4me3 determined in pro-B cells and binding sites of Pax5 and Ikaros identified in pre-B cells are shown.2,5,38,39 The UCSC Genome Browser was used to visualize binding patterns. Below, exon-intron structures of genes and a scale bar in kilobases are shown. Transcript direction is indicated by an arrow. The results of 2 ChIP-seq analyses are shown (Mef2c-I and Mef2c-II). (B) Frequency of Mef2c peaks with the indicated distance to the nearest TSS is shown. (C) Consensus sequences within 150 bp of the Mef2c-peak summits identified by de novo–motif discovery. The frequency of the motif around Mef2c peaks (n = 500) and the calculated E values are given. (D) The frequency of known E2a, Pax5, Ikaros, or Runx1 peaks2,5,38,39 within the indicated distance from Mef2c-peak summits is shown.
Genome-wide mapping of direct target genes of Mef2c identified by ChIP-seq analysis in pro-B cells. (A) Mef2c bound regions in 4 gene loci identified by peak calling. Known binding sites of E2a, Runx1, and regions with H3K4me3 determined in pro-B cells and binding sites of Pax5 and Ikaros identified in pre-B cells are shown.2,5,38,39 The UCSC Genome Browser was used to visualize binding patterns. Below, exon-intron structures of genes and a scale bar in kilobases are shown. Transcript direction is indicated by an arrow. The results of 2 ChIP-seq analyses are shown (Mef2c-I and Mef2c-II). (B) Frequency of Mef2c peaks with the indicated distance to the nearest TSS is shown. (C) Consensus sequences within 150 bp of the Mef2c-peak summits identified by de novo–motif discovery. The frequency of the motif around Mef2c peaks (n = 500) and the calculated E values are given. (D) The frequency of known E2a, Pax5, Ikaros, or Runx1 peaks2,5,38,39 within the indicated distance from Mef2c-peak summits is shown.
As expected, de novo–motif discovery programs identified a known Mef2c consensus recognition sequence in 89.4% of all random peaks analyzed (n = 500) (Figure 3C; supplemental Figure 3B). Moreover, motifs containing the consensus sequence for other known TFs, including Runx1, Ebf1, Ikaros,36 or members of the Ets factor family, were found at frequencies ranging from 18% to 32% (Figure 3C; supplemental Figure 3B). Motif discovery approaches37 identified consensus binding sites for Pax5 and E2a factors at frequencies of 8.1% and 7.0%, respectively (supplemental Figure 3B). Analyzing published E2a-, Runx1-, Pax5-, and Ikaros-binding regions observed in pro-B-2,5 or pre-B-cell lines,38,39 we observed coincidental binding of 19.6%, 17.5%, 13.0%, and 8.7% with Mef2c peaks, respectively, which was highly significant for all factors (Figure 3D).
The comparative analysis of our ChIP results suggested that Mef2c forms regulatory complexes with known TFs that orchestrate B-cell development. To investigate this further, we examined the binding site of Mef2c and other B-cell TFs generated by ENCODE40 in human GM12878 transformed peripheral B lymphocytes. This allows confirmation of coincident binding in a single cell type, albeit with a mature phenotype. Mef2c occupancy sites not only significantly coincided with the same B-cell TFs found in pro-B/pre-B cells (eg, Pax5, E2a, and Ebf1), but also demonstrated significant overlap with binding sites for Irf4 and Egr1 (Figure 4A-B). Furthermore, binding was observed in promoter/enhancer regions of several key pre-B-cell regulatory genes that are deregulated in Mef2c/d-deficient mice, including Irf4, Ikzf1, Hdac5/9, Zfp36, and Erg2 (Figure 4A). Coincident binding of 5 factors (Ebf1, E2a, Pax5, Irf4, and Egr1) was detected in 9% of all Mef2c peaks (Figure 4C). Hierarchial clustering and pairwise analysis using Bioconductor software packages41,42 confirmed the high significance of this finding (P < e12 000). The close proximity of the peak summits between Mef2c and several TFs suggests collaborative or perhaps even cooperative transcriptional regulation through protein-protein interactions (supplemental Figure 4A), a well-known attribute of Mef2 proteins.43-45 To pursue this hypothesis, coimmunoprecipitation analysis was performed in pre-B cells, confirming an interaction of Mef2c with Irf4 and Egr1 (Figure 4D; supplemental Figure 4B). Analysis of expression data generated by Gene Expression Commons22 revealed that increased transcription of Egr2 and Irf4 at the pre-B-cell stage coincides with the upregulation of Mef2c-target genes, suggesting that Egr and Irf4 collaborate with Mef2 proteins to induce or accelerate transcription of several pre-BCR regulatory genes (Figure 4E). Interestingly, Mef2c binding was also observed in the promoters of the genes encoding Hdac5 and Hdac9, which are known to repress Mef2c activity.46 Taken together, our results suggest that Mef2c is not only a critical regulator of IEG expression in pre-B cells, but also regulates the expression of transcriptional activators and repressors that likely collaborate with Mef2c and other B-cell TFs to drive differentiation.
Analysis of Mef2c complexes in GM12878-transformed peripheral B cells and murine pre-B cells. (A) Regulatory regions of putative Mef2c/d target genes occupied by Mef2c and the indicated TFs (light blue) are shown. ChIP-seq data sets were generated from ENCODE40 and visualized using the UCSC Genome Browser, showing conservation (100 species), exon-intron structures of genes, and a scale bar in kilobases. Transcript direction is indicated by an arrow. (B) The frequency of Irf4, Egr1, Ebf1, Pax5, and E2a peaks within the indicated distance from Mef2c summits in GM12878 B cells is depicted. (C) Hierarchical clustering of Mef2c-binding sites with coincident binding with Irf4, Egr1, Ebf1, Pax5, or E2a sites generated using GENE-E.41 For each peak (y-axis), green indicates overlapping bound regions (minimum of 2 bp). Peak regions for individual TFs were set at a standard width of 300 bp. (D) Interaction of Mef2c with Irf4 and Egr1 in Blnk-deficient pre-B cells. Mef2c was precipitated using specific antibody and protein G Sepharose beads. Irf4 or Egr1 were visualized using specific antibodies. Gapdh served as a control. Results shown are representative of 2 independent experiments performed. Protein molecular weights are given (kilodalton). (E) Gene expression profiles of Mef2c/d and other pre-B-cell regulatory genes during murine B-cell differentiation were collected from the Gene Expression Commons22 and are represented in a heat map. CLP, common lymphoid progenitor; pro-B cell (Hardy Fraction [Fr.] B); large pre-B cell (Fr.C′); small pre-B cell (Fr.D); immature B cell (Fr.E).
Analysis of Mef2c complexes in GM12878-transformed peripheral B cells and murine pre-B cells. (A) Regulatory regions of putative Mef2c/d target genes occupied by Mef2c and the indicated TFs (light blue) are shown. ChIP-seq data sets were generated from ENCODE40 and visualized using the UCSC Genome Browser, showing conservation (100 species), exon-intron structures of genes, and a scale bar in kilobases. Transcript direction is indicated by an arrow. (B) The frequency of Irf4, Egr1, Ebf1, Pax5, and E2a peaks within the indicated distance from Mef2c summits in GM12878 B cells is depicted. (C) Hierarchical clustering of Mef2c-binding sites with coincident binding with Irf4, Egr1, Ebf1, Pax5, or E2a sites generated using GENE-E.41 For each peak (y-axis), green indicates overlapping bound regions (minimum of 2 bp). Peak regions for individual TFs were set at a standard width of 300 bp. (D) Interaction of Mef2c with Irf4 and Egr1 in Blnk-deficient pre-B cells. Mef2c was precipitated using specific antibody and protein G Sepharose beads. Irf4 or Egr1 were visualized using specific antibodies. Gapdh served as a control. Results shown are representative of 2 independent experiments performed. Protein molecular weights are given (kilodalton). (E) Gene expression profiles of Mef2c/d and other pre-B-cell regulatory genes during murine B-cell differentiation were collected from the Gene Expression Commons22 and are represented in a heat map. CLP, common lymphoid progenitor; pro-B cell (Hardy Fraction [Fr.] B); large pre-B cell (Fr.C′); small pre-B cell (Fr.D); immature B cell (Fr.E).
Mef2c/d are phosphorylated after pre-BCR signaling
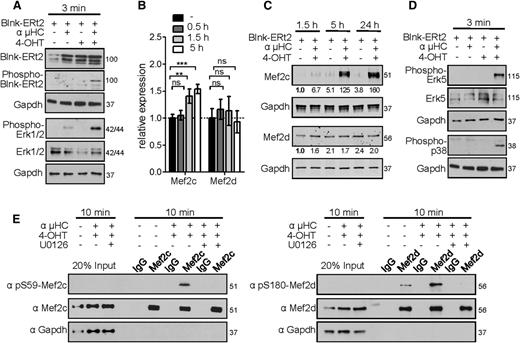
To investigate whether the transcriptional activity of Mef2c/d is directly linked to the pre-B-cell signaling pathway, we used a Blnk-deficient pre-B-cell line. A tamoxifen (4-OHT)-inducible Blnk was reintroduced into pre-B cells using a retroviral construct, in which Blnk is fused to a mutated ligand-binding domain of the estrogen receptor (Blnk-ERt2). This model system allows the induction of pre-BCR signaling in a temporally regulated manner.30,31 Stimulation of pre-BCR signaling using 4-OHT and anti-µHC-specific crosslinking antibody was confirmed by phosphorylation of Blnk and the MAPKs extracellular signal-regulated kinase 1/2 (Erk1/2) (Figure 5A). Erk1/2 has been shown to activate the cAMP response element-binding protein TF, and thereby stimulate transcription of Mef2c and Mef2d by ∼1.5-fold.47 In agreement, stimulation of pre-BCR led to minor upregulation of Mef2c (1.5-fold, P < .01), but no significant increase of Mef2d transcript levels was detected (Figure 5B). However, we detected a significant increase (up to 160-fold) of Mef2c protein level after pre-BCR stimulation for 1.5, 5, or 24 hours, whereas the amount of Mef2d protein remained nearly constant (Figure 5C). Increased protein levels after stimulation correlated with a significantly higher protein half-life of Mef2c, which in the nonstimulated stage is highly susceptible to degradation by the proteasome complex (supplemental Figure 5).
Mef2c/d proteins are phosphorylated after pre-BCR signaling. (A) Western blot analysis of Blnk-ERt2, phospho-Blnk-ERt2, phospho-Erk1/2, and Erk1/2 in Blnk-deficient pre-B cells transduced with Blnk-ERt2 and stimulated for 3 minutes with an IgH antibody (µHC) and tamoxifen (4-OHT) as indicated. Protein molecular weights are indicated in kilodaltons. (B) Relative transcript levels of Mef2c and Mef2d in pre-B cells stimulated for the indicated time periods identified by RNA-seq analysis and confirmed by RT-PCR. Error bars represent ± SD. **P < .01; ***P < .001; ns, not significant. (C) Immunoblot analysis and quantification of Mef2c and Mef2d proteins in Blnk-ERt2–overexpressing pre-B cells stimulated as indicated. Quantification was performed using the Odyssey Image scanner system (LI-COR Biosciences). Numbers indicate the level of Mef2c/d protein relative to that in the untreated control (first lane). Gapdh served as a loading control. (D) Western blot analysis of phospho-p38, phospho-Erk5, and Erk5 in Blnk-deficient pre-B cells as described in panel A. (E) Immunoprecipitation of Mef2c/d in Blnk-ERt2–overexpressing pre-B cells stimulated with IgH and 4-OHT for 10 minutes and with or without treatment with Mek inhibitor U0126. Phosphorylation of the Mef2 proteins at the indicated serines was visualized using specific antibodies. Gapdh served as a loading control. Twenty percent of the cell lysate was used as input control. Results shown are representative of at least 2 independent experiments performed. Protein molecular weights are indicated in kilodaltons.
Mef2c/d proteins are phosphorylated after pre-BCR signaling. (A) Western blot analysis of Blnk-ERt2, phospho-Blnk-ERt2, phospho-Erk1/2, and Erk1/2 in Blnk-deficient pre-B cells transduced with Blnk-ERt2 and stimulated for 3 minutes with an IgH antibody (µHC) and tamoxifen (4-OHT) as indicated. Protein molecular weights are indicated in kilodaltons. (B) Relative transcript levels of Mef2c and Mef2d in pre-B cells stimulated for the indicated time periods identified by RNA-seq analysis and confirmed by RT-PCR. Error bars represent ± SD. **P < .01; ***P < .001; ns, not significant. (C) Immunoblot analysis and quantification of Mef2c and Mef2d proteins in Blnk-ERt2–overexpressing pre-B cells stimulated as indicated. Quantification was performed using the Odyssey Image scanner system (LI-COR Biosciences). Numbers indicate the level of Mef2c/d protein relative to that in the untreated control (first lane). Gapdh served as a loading control. (D) Western blot analysis of phospho-p38, phospho-Erk5, and Erk5 in Blnk-deficient pre-B cells as described in panel A. (E) Immunoprecipitation of Mef2c/d in Blnk-ERt2–overexpressing pre-B cells stimulated with IgH and 4-OHT for 10 minutes and with or without treatment with Mek inhibitor U0126. Phosphorylation of the Mef2 proteins at the indicated serines was visualized using specific antibodies. Gapdh served as a loading control. Twenty percent of the cell lysate was used as input control. Results shown are representative of at least 2 independent experiments performed. Protein molecular weights are indicated in kilodaltons.
To further investigate the role of Mef2c/d during pre-BCR signaling, we sought to determine whether the MAPK p38 or Erk5 pathways, which are known to phosphorylate and activate Mef2c or both Mef2c and Mef2d, respectively,48,49 are activated by the pre-BCR. Mapk14 (encoding p38α) is expressed primarily in hematopoietic progenitors, whereas Mapk7 (encoding Erk5) and Mapk1 (encoding Erk2) show relatively high expression levels during early B-cell development.22 Congruently, simultaneous Blnk activation and µHC stimulation lead to Erk5 and p38 phosphorylation (Figure 5D). Although serine (S) 387 of Mef2c was constitutively phosphorylated, presumably through IL7 signaling (supplemental Figure 6), 10 minutes of pre-BCR stimulation resulted in highly increased phosphorylation of Mef2c at S59 and of Mef2d at S180 (Figure 5E). Whereas S387 phosphorylation has been shown to be mediated by Erk5,49 phosphorylation of S59-Mef2c is likely mediated by activation of the protein kinase CK2 via activation of Erk, leading to greatly increased DNA-binding affinity.50-53 Consistent with the observed phosphorylation being mediated by Erk5, phosphorylation of Mef2c and Mef2d could be blocked by the Mek inhibitor U0126 (Figure 5E).
Activation of Mef2c/d target gene expression during pre-BCR signaling
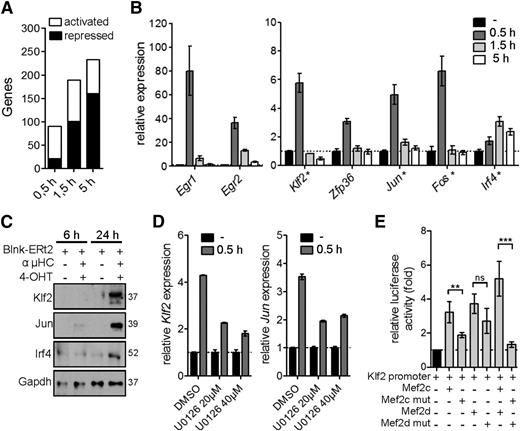
In a next step toward defining the role of Mef2c/d in modulating the gene expression program after pre-BCR signaling, we performed a genome-wide transcript analysis at different time periods after pre-BCR signaling in the tamoxifen-inducible Blnk culture model. We identified 90, 189, and 233 deregulated genes (≥2-fold) after 0.5 hours, 1.5 hours, and 5 hours of pre-BCR stimulation, respectively (Figure 6A). Significantly, 14.5% of the transiently upregulated genes at 0.5 hours were deregulated in Mef2c/d-deficient pro-B cells (≥1.5-fold, P ≤ .05) (Figure 6B). The induction of Klf2, Jun, and Irf4 gene expression could be confirmed at the protein level (Figure 6C). Furthermore, inhibition with the Mek inhibitor U0126 during pre-BCR stimulation led to significantly reduced induction of the Mef2c-target genes Klf2 and Jun (Figure 6D).
Activation of Mef2c/d target gene expression during pre-BCR signaling. (A) Activated and repressed gene expression (≥2-fold; identified by RNA-seq analysis) in Blnk-ERt2–expressing pre-B cells stimulated with IgH antibody and tamoxifen (4-OHT) for indicated time periods. (B) Relative transcript levels of a selection of genes (Egr1/2, Klf2, Zfp36, Jun, Fos, Irf4) in Blnk-ERt2+ pre-B cells stimulated for different time points using IgH and 4-OHT. qRT-PCR was used to confirm transcript level of a subset of genes, indicated by a star (★). (C) Immunoblot analysis of Klf2, Jun, and Irf4 in Blnk-ERt2–overexpressing cells stimulated for 6 and 24 hours with IgH and 4-OHT. Gapdh served as a control. (D) Relative expression of Klf2 and Jun in Blnk-ERt2+ pre-B cells stimulated with IgH and 4-OHT for 30 minutes and treated with different concentrations of Mek inhibitor U0126 as indicated. (E) Analysis of the impact of Mef2c and Mef2d mutants on Klf2 expression using a reporter gene assay. Cos-7 cells were transfected with a luciferase expression plasmid containing a Klf2 promotor with a Mef2c/d wild-type expression plasmid or Mef2c (S59A, T293A, T300A, S387A) or Mef2d (S180A) phosphorylation mutants or empty control plasmid. Firefly luciferase activity (relative to renilla luciferase) was determined. **P < .01; ***P < .001; ns, not significant. Results shown are representative of 2 independent experiments performed.
Activation of Mef2c/d target gene expression during pre-BCR signaling. (A) Activated and repressed gene expression (≥2-fold; identified by RNA-seq analysis) in Blnk-ERt2–expressing pre-B cells stimulated with IgH antibody and tamoxifen (4-OHT) for indicated time periods. (B) Relative transcript levels of a selection of genes (Egr1/2, Klf2, Zfp36, Jun, Fos, Irf4) in Blnk-ERt2+ pre-B cells stimulated for different time points using IgH and 4-OHT. qRT-PCR was used to confirm transcript level of a subset of genes, indicated by a star (★). (C) Immunoblot analysis of Klf2, Jun, and Irf4 in Blnk-ERt2–overexpressing cells stimulated for 6 and 24 hours with IgH and 4-OHT. Gapdh served as a control. (D) Relative expression of Klf2 and Jun in Blnk-ERt2+ pre-B cells stimulated with IgH and 4-OHT for 30 minutes and treated with different concentrations of Mek inhibitor U0126 as indicated. (E) Analysis of the impact of Mef2c and Mef2d mutants on Klf2 expression using a reporter gene assay. Cos-7 cells were transfected with a luciferase expression plasmid containing a Klf2 promotor with a Mef2c/d wild-type expression plasmid or Mef2c (S59A, T293A, T300A, S387A) or Mef2d (S180A) phosphorylation mutants or empty control plasmid. Firefly luciferase activity (relative to renilla luciferase) was determined. **P < .01; ***P < .001; ns, not significant. Results shown are representative of 2 independent experiments performed.
To further investigate the influence of phosphorylation of Mef2c/d on TF activity, we performed reporter gene assays using a Klf2 reporter gene construct containing a Mef2-binding site and wild-type or phosphorylation mutants of Mef2c and Mef2d (Figure 6E). Expression of either Mef2c or Mef2d led to increased luciferase activity, whereas substitution with Mef2c/d phosphorylation mutants resulted in strongly decreased luciferase activity (Figure 6E). Taken together, our data are consistent with the interpretation that not only the classical IEG family genes but also Klf2 and Irf4 are stimulated via phosphorylation and activation of Mef2 proteins by the Erk pathway during pre-BCR signaling.
Genome-wide identification of Klf2 target genes
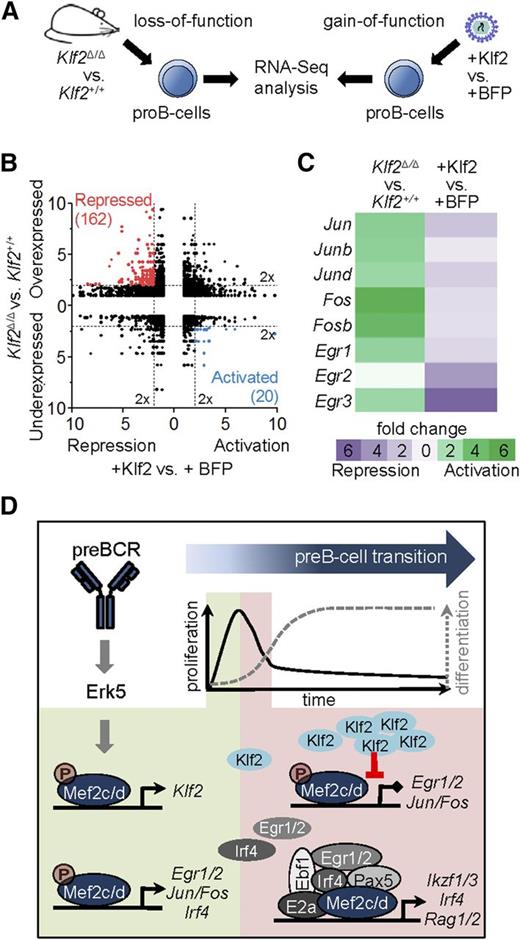
To achieve tight control of IEGs by the MAPK pathway, a number of pathways are simultaneously activated that serve to shutdown IEG expression by a variety of different mechanisms.54 To determine whether Mef2c-mediated activation of Klf2 serves as a negative regulator of IEG expression, we examined genome-wide transcript expression in gain-of-function and loss-of-function experiments (Figure 7A). For the first set, pro-B cells were engineered to constitutively express Klf2 or a control vector by retroviral transduction. These results were compared with transcriptome analysis of Klf2-deficient pro-B cells of Klf2Δ/ΔCd79ahCre/+ mice (Klf2Δ/Δ) and control mice (Cd79ahCre/+). By using stringent criteria for differential expression (2-fold differential expression, false discovery rate < 0.05), 182 Klf2 targets with reciprocal expression between the 2 approaches were identified (Figure 7B). Genes encoding AP1- and Egr-family members, including those identified as direct Mef2c targets during pre-BCR signaling (Figures 3A and 6B), were repressed in the gain-of-function model and activated in Klf2-deficient pro-B cells (Figure 7C). These results support the hypothesis that Klf2, a direct target of Mef2c, is an important negative regulator of IEG genes during pre-BCR signaling.
Genome-wide identification of Klf2 target genes. (A) Experimental setup for transcriptome analysis of Klf2 gain-of-function vs loss-of-function. For gain-of-function analysis, pro-B cells were transduced with retroviral vectors expressing blue fluorescent protein (BFP) with (+Klf2) or without Klf2 and sorted. For loss-of-function analysis, pro-B cells (Lin−CD19+B220medCD43hiIgL−) of Klf2Δ/ΔCd79ahCre/+ mice (Klf2Δ/Δ) or control mice (Cd79ahCre/+) were sorted by flow cytometry, RNA was isolated and sequenced. (B) Reciprocal deregulation of genes in loss-of-function (Klf2Δ/Δ vs Klf2+/+) and gain-of-function (+Klf2 vs +BFP) experiments in pro-B cells shown as fold change. Colors indicate 20 activated Klf2 target genes (blue) and 162 repressed Klf2 target genes (red). (C) Gene expression changes shown as fold change of genes encoding for AP1 family members (Jun/b/d, Fos/b) and early response genes (Egr1-3, Ier2) in Klf2 loss-of-function and gain-of-function experiments. (D) Schematic representation of the role of Mef2c/d proteins upon pre-BCR stimulation. Phosphorylation of Mef2c/d by Erk5 via pre-BCR signaling activates several transcriptional circuits. Mef2c/d activates the expression of Klf2 and IEGs (Egr1/2, Ier2, Jun/Fos). Klf2 suppresses IEG expression and thereby blocks pre-BCR–induced cell proliferation (negative feed-forward loop). In a positive feed-forward loop, Mef2c activates Egr and Irf4, the gene products of which collaborate with Mef2c and several key pre-B-cell regulatory factors to drive pre-B-cell differentiation.
Genome-wide identification of Klf2 target genes. (A) Experimental setup for transcriptome analysis of Klf2 gain-of-function vs loss-of-function. For gain-of-function analysis, pro-B cells were transduced with retroviral vectors expressing blue fluorescent protein (BFP) with (+Klf2) or without Klf2 and sorted. For loss-of-function analysis, pro-B cells (Lin−CD19+B220medCD43hiIgL−) of Klf2Δ/ΔCd79ahCre/+ mice (Klf2Δ/Δ) or control mice (Cd79ahCre/+) were sorted by flow cytometry, RNA was isolated and sequenced. (B) Reciprocal deregulation of genes in loss-of-function (Klf2Δ/Δ vs Klf2+/+) and gain-of-function (+Klf2 vs +BFP) experiments in pro-B cells shown as fold change. Colors indicate 20 activated Klf2 target genes (blue) and 162 repressed Klf2 target genes (red). (C) Gene expression changes shown as fold change of genes encoding for AP1 family members (Jun/b/d, Fos/b) and early response genes (Egr1-3, Ier2) in Klf2 loss-of-function and gain-of-function experiments. (D) Schematic representation of the role of Mef2c/d proteins upon pre-BCR stimulation. Phosphorylation of Mef2c/d by Erk5 via pre-BCR signaling activates several transcriptional circuits. Mef2c/d activates the expression of Klf2 and IEGs (Egr1/2, Ier2, Jun/Fos). Klf2 suppresses IEG expression and thereby blocks pre-BCR–induced cell proliferation (negative feed-forward loop). In a positive feed-forward loop, Mef2c activates Egr and Irf4, the gene products of which collaborate with Mef2c and several key pre-B-cell regulatory factors to drive pre-B-cell differentiation.
Discussion
The pre-B-cell stage is a critical checkpoint in B-cell development. Signals initiated by the pre-BCR through PI3K and MAPKs trigger, in cooperation with IL7 receptor signals, a signaling cascade that induces a short proliferation pulse of large pre-B cells, transition to a resting state, induction of IgL gene rearrangements, and further differentiation.7,9,55 In this study, we could show that the MADS proteins Mef2c and Mef2d are critical integrators of pre-BCR signaling, translating the phosphorylation events triggered by the MAPK cascade to transcriptional regulators that orchestrate the transition to the immature B-cell stage through a circuitry of positive and negative feed-forward loops (Figure 7D). In addition to well-characterized IEGs that initiate cell proliferation and differentiation programs, our study has identified other key regulatory pre-B-cell genes (eg, Klf2, Hdac5, and Irf4) that are likely targets of Mef2c/d through combinatorial interactions with B-cell TFs. In the absence of Mef2c/d TFs, B-cell development is blocked at the pre-B-cell stage.
Mechanistically, an important finding is the demonstration that the MAPK Erk5 is activated upon pre-BCR stimulation and mediates phosphorylation of Mef2c/d. The significance of the signaling pathway leading from Ras GTPase to the MAPK cascade in pre-BCR proliferation and differentiation has long been recognized and attributed to the Erk1/2 cascade.47,56,57 However, these studies did not investigate the Erk5 pathway, which is the fourth and most recently identified MAPK cascade. Erk5 shares many characteristics of Erk1/2, including a Thr-Glu-Tyr (TEY) activation motif, responsiveness to Ras, and sensitivity to the same chemical inhibitors (eg, PD98059 and U0126),58,59 but is unique in its ability to directly activate Mef2c/d proteins. Hence, our data demonstrate that the Erk5 pathway may be as important as the Erk1/2 cascade for relaying pre-BCR signals; a hypothesis strengthened by the distinct expression pattern of Erk5 in the lymphoid lineage and that provides a model of how specificity in gene regulation can be achieved by the MAPK pathways.
Activation of MADS proteins through the MAPK signal cascade is a highly conserved pathway by which cells respond to extracellular signals and is best studied for the serum response factor (SRF), a MADS protein lacking the Mef2 domain.20,60,61 Notably, SRF is essential for positive selection of thymocytes,62,63 in analogy to the essential role revealed here for Mef2c/d in early B-cell development. However, in contrast to SRF, for which it is established that transcriptional activation by MAPK is mediated through collaborating TFs, Mef2c/d proteins are activated directly through phosphorylation of specific residues by MAPK Erk5, leading to an increase in their transcriptional activity.64-66 We were able to confirm this for pre-B cells using Mef2c mutants. Interestingly, we also observed that Mef2c but not Mef2d protein levels significantly increased after pre-BCR stimulation, which could mechanistically be traced to increased protein stability, although an additional impact on translational controls cannot be excluded.67,68
As mediator of the MAPK response, it is not surprising that many of the Mef2c target genes identified in our study have been characterized as IEGs, also known as the “gateway to the genetic response.” These regulatory genes form circuits with recurring network motifs, which include positive and negative feed-forward loops, insuring both a robust response to the stimulus but also tight control to prevent uncontrolled proliferation.54,69 In addition to IEGs encoding AP1- and Egr-family members, which are likely involved in the control of both cell cycle and further differentiation,70,71 our analysis also identified 2 negative regulators of early IEGs whose expression is directly controlled by Mef2c. Zfp36 is an RNA-binding protein that promotes degradation of mRNAs of several components of the AP1 family,54,72 whereas Klf2 is a TF implicated in both proliferation and quiescence of lymphoid cells.73 To confirm a repressor role of Klf2 at the pre-BCR checkpoint, we identified direct target genes of Klf2 by analysis of pro-/pre-B cells in Klf2−/− mice. We could confirm a critical role of Klf2 in the suppression of IEG genes, which would be predicted to attenuate the proliferation stimulus. In agreement with our results, Klf2 has recently been shown to inhibit pre-BCR-mediated proliferation.74 Thus, Mef2c not only activates AP1/Egr genes, but also represses them by activating the Klf2 repressor (Figure 7D). Importantly, this “incoherent feed-forward loop” motif, in which 2 arms of a network act in opposition, is a key network motif for pulse dynamics.69
Lastly, the results presented here support the hypothesis that Mef2c/d play important roles in early B-cell development by forming multicomponent regulatory complexes with several B-cell TFs. This hypothesis is based on analysis of Mef2c-binding studies in pro-B- and pre-B cells, which demonstrated that 13% to 20% of the known E2a, Ikaros/Aiolos, and Pax5 occupancy sites showed coincident binding with Mef2c. Ikaros and Pax5 are known to drive the differentiation of B-cell progenitors by activating Irf4 and Rag1 gene expression.38,39 These 2 genes are also deregulated in Mef2c/d knockouts, thus it is likely that Mef2c coregulates these genes at the pre-B-cell stage. In confirmation and extension of our hypothesis, analysis of genome-wide occupancy data generated from human B lymphocytes showed that close to 10% of the Mef2c peaks bound to gene regulatory regions also occupied by 5 TFs: E2a, Ebf1, Pax5, Egr1, and Irf4. As the latter 2 factors are encoded by potential direct target genes of Mef2c, these results reveal a coherent feed-forward loop for driving B-cell differentiation (Figure 7D). Our work also confirmed physical interactions of Mefc with Irf4 and Egr1, supporting the idea that such combinatorial associations may establish a unique transcription code, a known attribute of Mef2 proteins.43-45
In summary, this study revealed an essential role for the Mef2c/d proteins in regulating transition through the first checkpoint in early development by integrating the MAPK signals of the pre-BCR to a downstream transcriptional network, thereby controlling proliferation and orchestrating the progression through the next phase of B-cell development. In view of growing recognition that disruption of pre-BCR controls can lead to leukemia,75,76 this work provides insight into the mechanism by which MEF2C and MEF2D disruption23-25 contributes to acute lymphoblastic leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Düsedau and J. Hennesen for sorting cells by flow cytometry, G. Pilnitz-Stolze for histologic preparation, E. N. Olson for providing Mef2cfl/fl and Mef2d−/− animals, M. Reth for the Cd79ahCre/+ mice, and H. Jumaa for the Blnk-deficient pre-B-cell line.
This work was supported by the Else Kröner-Fresenius-Stiftung (2010/A106), Deutsche José Carreras Leukämie-Stiftung e.V. (11/28f), Deutsche Krebshilfe (#109528) (C.S.) and the Deutsche Forschungsgemeinschaft (GK1660, TRR130) (H.-M.J.). The Heinrich-Pette-Institute, Leibnitz Institute for Experimental Virology is financed by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
Authorship
Contribution: L.U., F.H., M.F., N.K., D.I., M.Z., and U.M. performed research; M.A. and M.S. performed bioinformatic analysis; J.H. designed experiments, performed research, and analyzed data; W.S. and H.-M.J. discussed results and the manuscript; C.S. designed the study, supervised work, and analyzed data; and J.H. and C.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.U. is Division of Cell and Gene Therapy, Bone Marrow Transplantation Unit, Center for Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Correspondence: Carol Stocking, Heinrich-Pette-Institute, Martinistrasse 52, 20251 Hamburg, Germany; e-mail: carol.stocking@hpi.uni-hamburg.de.

![Figure 1. Functional deletion of Mef2c/d in B-cell progenitors leads to a developmental block of B-cell differentiation. (A) Flow cytometric analysis to detect and enumerate pro-B cells (Lin−CD19+B220medCD43hiIgL−), pre-B cells (Lin−CD19+B220medCD43med/loIgL−), immature B cells (Lin−CD19+B220medCD43med/loIgL+), and mature (recirculating) B cells (Lin−CD19+B220hiCD43−IgL+) in the BM of Mef2cΔ/Δ (light blue), Mef2dΔ/Δ (green), Mef2c/dΔ/Δ (red), and control mice (Mef2c/d+/+, dark blue). Pre-B cells were separated by forward light scatter (FSC) into large and small pre-B cells, as indicated. Representative flow cytometry plots of Mef2c/dΔ/Δ and Mef2c/d+/+ mice are shown (top). Graph (bottom) depicts the number of different B-cell subtypes within total BM. Each mouse is represented by a dot (n ≥ 8 per genotype). Horizontal lines indicate the median. **P < .01; ***P < .001; ns, not significant. (B-C) Evaluation of B cells in blood (n = 5 per cohort) (B) and spleen (n = 12 per cohort) (C) of Mef2c/dΔ/Δ and control mice (Mef2c/d+/+) using FACS analysis. Graph depicts the percentage of CD19+B220+ B cells. Dots represent individual mice and bars indicate the median. (D) B220/CD45R staining of B-cell zones (indicated by a star [★]) from lymph nodes of Mef2c/dΔ/Δ and Mef2c/d+/+ mice. B-cell zones are lacking in lymph nodes of Mef2c/dΔ/Δ mice. Representative stainings from 2 experiments (I and II) using 2 mice per group are shown. Bar, 200 µm. (E) Schematic representation of the observed B-cell differentiation block at the pre-B-cell stage in BM of Mef2c/dΔ/Δ mice. The stage in B-cell development at which Cd79a-hCre–mediated excision occurs is indicated. Receptors that are important for the proliferative expansion of the specific B-cell types are depicted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/5/10.1182_blood-2015-04-643270/4/m_572f1.jpeg?Expires=1765891610&Signature=NWo-Id2qhL7ywtpDdbSmxTtAkn3WdlCSFZjVrJnJihq15AWd2HmQqL~QpjWrvXhm5DjyPFyhKJ7H8jcGpqMptNIqYqnz~y5~m19bk~PVnkDY9sDTEeXRUDpXINnDzV0s~2o7nbLzeqa39e5Vk-80w6560Y3SktsR3uGHDY~VVamEvsAcvezHjbayiRtm9ilqV8jbzOkLpGD1wSPAnF9xGtmOR6~rQM-iDik3c7NZAvQOWDpPIOpgtsG2M-ZHQCmrfGBNgL5PTs~anp2guI4W5roVDo5VsB~7WWZm~xPot5pCF5s26LcUH1xexp3uw8I2T0xiknxjXgXzGVsSg0mN3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Analysis of Mef2c complexes in GM12878-transformed peripheral B cells and murine pre-B cells. (A) Regulatory regions of putative Mef2c/d target genes occupied by Mef2c and the indicated TFs (light blue) are shown. ChIP-seq data sets were generated from ENCODE40 and visualized using the UCSC Genome Browser, showing conservation (100 species), exon-intron structures of genes, and a scale bar in kilobases. Transcript direction is indicated by an arrow. (B) The frequency of Irf4, Egr1, Ebf1, Pax5, and E2a peaks within the indicated distance from Mef2c summits in GM12878 B cells is depicted. (C) Hierarchical clustering of Mef2c-binding sites with coincident binding with Irf4, Egr1, Ebf1, Pax5, or E2a sites generated using GENE-E.41 For each peak (y-axis), green indicates overlapping bound regions (minimum of 2 bp). Peak regions for individual TFs were set at a standard width of 300 bp. (D) Interaction of Mef2c with Irf4 and Egr1 in Blnk-deficient pre-B cells. Mef2c was precipitated using specific antibody and protein G Sepharose beads. Irf4 or Egr1 were visualized using specific antibodies. Gapdh served as a control. Results shown are representative of 2 independent experiments performed. Protein molecular weights are given (kilodalton). (E) Gene expression profiles of Mef2c/d and other pre-B-cell regulatory genes during murine B-cell differentiation were collected from the Gene Expression Commons22 and are represented in a heat map. CLP, common lymphoid progenitor; pro-B cell (Hardy Fraction [Fr.] B); large pre-B cell (Fr.C′); small pre-B cell (Fr.D); immature B cell (Fr.E).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/5/10.1182_blood-2015-04-643270/4/m_572f4.jpeg?Expires=1765891610&Signature=1tddLPBD7asXHMZUzrKXant7KcYdvpoEvo643tW7z9tKBNtVEyg4-g3PyDDWrW0Y5u~EqrHRWs2~vrZjBjRtxygsLL4KdPoSsneCi9AaPUvD5~xyWoyFjUXPz8l-KOp99E4Jp~2zlpqIEvqUo6B~hykhXPdwRrkkSyuJvXT7g05uVYFPI8~yGXchyoIdVJsf2kpxRCxExxfqheE9lMOb9j7yZu0j7cQf7kUOusbatLP2qPvwYnrzdjR4CFM8pY~q7w5mx~Lo-psGUNKbHMZxKnLl5nBsdpQ6z4YycxMg53w-mAsHhNxmxxv9sjwPKWHwm9vNYDXQqHT1P7plAF36Vw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)