Key Points
Exome sequencing of severe hemophilia A patients with/without inhibitors identified rare, damaging variants in immunoregulatory genes.
Replication confirmed the association of rs3754689 in a conserved haplotype region surrounding the LCT locus with inhibitor development.
Abstract
The development of neutralizing antibodies (inhibitors) against coagulation factor VIII (FVIII) is the most problematic and costly complication of FVIII replacement therapy that affects up to 30% of previously untreated patients with severe hemophilia A. The development of inhibitors is a multifactorial complication involving environmental and genetic factors. Among the latter, F8 gene mutations, ethnicity, family history of inhibitors, and polymorphisms affecting genes involved in the immune response have been previously investigated. To identify novel genetic elements underling the risk of inhibitor development in patients with severe hemophilia A, we applied whole-exome sequencing (WES) and data analysis in a selected group of 26 Italian patients with (n = 17) and without (n = 9) inhibitors. WES revealed several rare, damaging variants in immunoregulatory genes as novel candidate mutations. A case-control association analysis using Cochran-Armitage and Fisher’s exact statistical tests identified 1364 statistically significant variants. Hierarchical clustering of these genetic variants showed 2 distinct patterns of homozygous variants with a protective or harmful role in inhibitor development. When looking solely at coding variants, a total of 28 nonsynonymous variants were identified and replicated in 53 inhibitor-positive and 174 inhibitor-negative Italian severe hemophilia A patients using a TaqMan genotyping assay. The genotyping results revealed 10 variants showing estimated odds ratios in the same direction as in the discovery phase and confirmed the association of the rs3754689 missense variant (OR 0.58; 95% CI 0.36-0.94; P = .028) in a highly conserved haplotype region surrounding the LCT locus on chromosome 2q21 with inhibitor development.
Introduction
Hemophilia A (HA) is an inherited bleeding disorder characterized by factor VIII (FVIII) deficiency due to mutations in the F8 gene.1 HA treatment consists of replacement with FVIII concentrates and is highly effective in stopping and preventing hemorrhage. However, up to 30% of previously untreated patients with severe HA develop anti-FVIII neutralizing alloantibodies (referred to as inhibitors), which impair the efficacy of replacement therapy and render the management of bleeds difficult.2 For these reasons, inhibitor development is the most problematic and costly complication of FVIII replacement therapy. To purge the immune system of alloantibodies against FVIII, the immune tolerance induction strategy is frequently implemented. However, it is a demanding and costly treatment that involves long-term regular infusions of high doses of FVIII with a success rate of 60% to 80%.3-5 Therefore, prevention of inhibitor development is an urgent and important issue that requires a deeper insight into the etiology of this complication.
It has been postulated that inhibitor formation results from a complex interaction of environmental and genetic factors.6-8 Studies on genetic determinants of inhibitor development have indicated numerous genetic markers, such as the type of causative F8 gene mutation, single nucleotide polymorphisms (SNPs) in the HLA locus and in other immunoregulatory genes, ethnic background, and family history of inhibitors, to be involved in the etiology of this treatment complication.9 Recently, Gouw et al10 performed a systematic review and meta-analysis to associate the type of F8 gene mutation with inhibitor development in severe HA patients and found null mutations that lead to a complete lack of protein expression to be the strongest risk factors for inhibitor development. Studies on genetic determinants other than F8 mutations identified SNPs in genomic regions associated with immunoregulatory genes.9 These include the HLA class II molecules,11-14 which have an essential role in the presentation of FVIII peptides to CD4+ T-helper cells and pro- and anti-inflammatory cytokines such as interleukin (IL)-1, IL-2, IL-10, CTLA4, and tumor necrosis factor α.13,15-19 Moreover, polymorphism p.R131H in the Fc Fragment of immunoglobulin (Ig)G (FCGR2A) encoding Fc γ receptor 2A expressed on immune cells has recently been associated with inhibitor development.20 However, due to the poor reproducibility of the genotyping results, small replication cohorts, or different genetic background of the examined populations, the gene targets that cause this predisposition are still largely unknown.
To identify novel genetic risk variants underlying inhibitor development, we applied whole-exome sequencing (WES) analysis in a group of Italian patients with severe HA, followed by replication of our results in a larger cohort of severe HA patients with and without inhibitors. This manuscript reports our experience using exome sequencing to discover novel, coding variants in immunoregulatory genes responsible for the development of inhibitory anti-FVIII alloantibodies in severe HA patients.
Materials and methods
Patients
Patients were selected from a cohort of 256 severe HA patients regularly followed up between 1960 and 2010 at the Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Fondazione Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Ca’ Granda, Ospedale Maggiore Policlinico (Milan, Italy). For the discovery phase involving exome sequencing, 26 Italian patients with severe HA were chosen, 17 with and 9 without a history of inhibitors. To reduce heterogeneity due to the F8 gene mutations, all patients chosen for the discovery phase carried intronic inversion mutations (n = 24 intron 22 inversions and n = 2 intron 1 inversions). Four brother pairs, including 2 discordant (E835-E1309 and E172-E1355) and 2 concordant E287-E1147 (inhibitor negative) and E1361-E502 (inhibitor positive) for inhibitor development, were included in the discovery phase. Clinical features of 26 Italian patients with severe HA are presented in supplemental Table 1, available on the Blood Web site. WES was performed at the Human Genome Sequencing Center (HGSC), Baylor College of Medicine (Houston, TX).
The replication study was conducted in 227 severe HA patients with (n = 53) and without (n = 174) inhibitor history. Patients with different types of the F8 gene mutations, such as intron 22 and 1 inversion mutations, missense, large deletions, frameshifts, and STOP gain, were chosen for replication. Nearly all patients (97.4%) were of Italian origin. Genotyping was performed in our center using a TaqMan SNP genotyping assay. A positive history of inhibitor development was defined as a positive Bethesda assay >0.5 Bethesda units (BU)/mL in 2 independent measurements. High titer inhibitor patients, henceforth called high responders (HR), were defined as those with inhibitor peak titers >5 BU/mL. All patients signed informed consent, and the study was approved by the Ethics Committee of the Fondazione IRCCS Ca’ Granda and was carried out in accordance with the Declaration of Helsinki.
Sequencing and data analysis
DNA isolation and preparation methods have been described in detail elsewhere.21 Individual exomes were enriched by the SeqCap EZ HGSC VCRome kit (Roche NimbleGen), a clinical research exome oligonucleotide panel designed to capture 189 028 nonoverlapping exons in 23 585 human genes, producing a total target size of 45.1 Mb. Sequencing of paired-end fragments was conducted on an Illumina HiSeq 2000 sequencing platform (Illumina, San Diego, CA) at the HGSC.
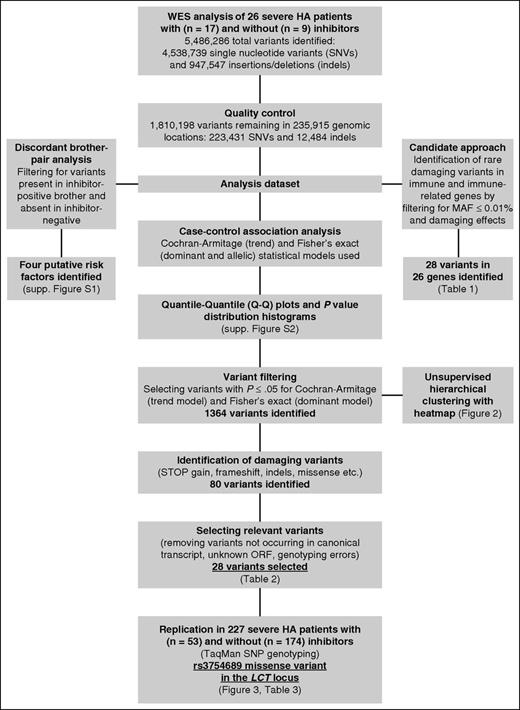
Data analysis on raw reads to produce individual variant call format (VCF)22 files was performed at HGSC. Raw reads were aligned to the reference human genome (hg19/GRCh37) and variants were called using the HGSC proprietary software 'Atlas2' (https://www.hgsc.bcm.edu/software/atlas-2). After variant calling, individual VCF files were merged using VCFtools,22 and variants not meeting quality control (QC) criteria, as specified by the HGSC, were removed: single nucleotide variant (SNV) posterior probability ≥0.95, number ≥3, variant-to-read ratio ≥0.1, variant reads mapping to different strand direction, and total coverage ≥6. Annotation of all QC-filtered variants was then conducted using the Variant Effect Predictor,23 including annotation with dbSNPvs138,24 ClinVar,25 Sorting Intolerant From Tolerant (SIFT),26 and Polyphen-2.27 We also obtained the minor allele frequencies (MAFs) from the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/) and the 1000 Genomes Project phase 3 populations.28,29 This merged, QC-filtered, and annotated VCF containing all variants called in 26 Italian patients with severe HA became the investigation dataset used in 3 different types of analysis: case-control association analysis, candidate approach to identify rare, damaging variants in immunoregulatory genes, and discordant brother-pair analysis (Figure 1).
Data analysis and filtering steps of genetic variants identified in WES of 26 severe hemophilia A patients. The analysis dataset was obtained by merging all annotated variants called in 26 Italian severe HA patients into a single dataset that was subsequently used in 3 different types of analysis: case-control association analysis, candidate approach to identify rare, damaging variants in immunoregulatory genes, and discordant brother-pair analysis.
Data analysis and filtering steps of genetic variants identified in WES of 26 severe hemophilia A patients. The analysis dataset was obtained by merging all annotated variants called in 26 Italian severe HA patients into a single dataset that was subsequently used in 3 different types of analysis: case-control association analysis, candidate approach to identify rare, damaging variants in immunoregulatory genes, and discordant brother-pair analysis.
A case-control association analysis was conducted between inhibitor-positive and inhibitor-negative patients using SnpSift case-control,30 taking into account 3 different genetic testing models: trend model (Cochran-Armitage test), a dominant model, and a comparison of raw allelic counts (both Fisher’s exact tests). Quantile-quantile (Q-Q) plots and P value distribution histograms were generated using custom scripts in R (R release 3.1.1; R Foundation for Statistical Computing, Vienna, Austria)31 on each derived set of P values. To obtain variants with putative functional impact, we only focused on the following variant types, henceforth referred to as damaging: STOP gains/losses, coding region insertions/deletions (indels), missense (not restricted to SIFT and Polyphen-2 predictions), and variants located in splice acceptor/donor sites.
Relatedness between individuals based on all variants that passed QC was estimated according to the Ajk statistics,32 with subsequent values plotted using custom R31 scripts. Unsupervised hierarchical clustering with heatmaps was performed by encoding variants as 0 (reference sequence, black), 1 (heterozygous variants, green), and 2 (homozygous variants, red) and then using R.31
Because WES was performed on Italian HA patients, we decided to use the MAFs derived from the Italian Tuscany (TSI) population from the 1000 Genomes Project, phase 3,28,29 as normal healthy Italian controls. Where the TSI MAF was not available, those from the HapMap phase 3 project33 population of Utah residents with Northern and Western European ancestry or the CSAgilent population of European descent from the ClinSeq project34 was used. To assess the association of each variant with inhibitor development, odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using R.31 The post hoc replication population’s power to confirm the associations found in the discovery phase was calculated using G*Power software, version 3.0.10,35 based on frequency and effect size of the identified variants, the replication sample size, and assuming a 0.05 2-tailed α error.
The candidate approach to identify rare, damaging variants in immunoregulatory genes was based on filtering all variants on MAF (both EVS and 1000 Genomes) <1%, followed by the identification of damaging variants predicted by both SIFT and Polyphen-2 as deleterious. The discordant brother-pair analysis is described in detail in supplemental Figure 1.
Replication and data analysis
A cohort of 227 severe HA patients with (n = 53) or without (n = 174) inhibitor history was subjected to TaqMan SNP genotyping replication (Thermo Fisher Scientific, Carlsbad, CA) using a StepOnePlus Real-Time Polymerase Chain Reaction system (Applied Biosystems) as described in the manufacturer’s instructions. Variants that showed a direction of association consistent with the discovery phase were further analyzed using logistic regression, adjusting for the relatedness among first-degree patients. The P values were 2 sided, and P < .05 was considered statistically significant. Multiple testing was calculated by Benjamini and Hochberg’s false discovery rate.36 To assess the susceptibility of all replicated variants for inhibitor development, ORs and 95% CIs were calculated. Data analysis was performed using Plink (v1.0737 ) and R.31
Results
We performed WES and data analysis that revealed a total of 5 486 286 variants (mean, 211 011) called across all samples, including 4 538 739 SNVs and 947 547 insertions/deletions (indels). After filtering out variants that did not pass QC in ≥1 patient, 1 810 198 variants (mean, 69 623) were identified in 235 915 unique genomic locations. These included 223 431 SNVs and 12 484 indels that were sequenced with an average read depth over each site of 59 (supplemental Table 2A). Filtering of these variants for novelty, ie, those that were not listed in dbSNPvs138 or any other variant database, we identified 28 510 variants, of which 25 945 were SNVs and 2565 were indels. Moreover, we identified 91 684 variants that were considered rare (MAF ≤1%), 14 619 low-frequency variants (MAF, 1-5%), and 129 612 common variants (MAF >5%) in the European-American population in the EVS and the combined European population in the 1000 Genomes Project.
Candidate approach to identify rare, damaging variants in immunoregulatory genes
To identify novel candidate mutations in immunoregulatory genes that could play a role in inhibitor development, we filtered rare variants for those with potential damaging consequences and missense variants predicted by both SIFT and Polyphen-2 as deleterious. In this way, we obtained a list of 2099 variants from which we selected 28 variants in 26 different genes that belonged to immunoglobulin and immunoglobulin-like genes, HLA loci, receptor and ligand chemokine subunits, interleukins, and other immune system modulators, as well as genes involved in DNA repair mechanisms that play a role in V(D)J recombination (Table 1). A total of 17 variants were identified as risk (found only in inhibitor-positive patients) and 11 as protective (found only in inhibitor-negative patients). Among the 28 SNVs, we found 3 novel variants: 2 missense variants in the CD83 gene and in the CD109 gene and 1 frameshift in the chemokine (C-X3-C motif) receptor 1 (CX3CR1) gene (Table 1). Moreover, we identified frameshift mutation c.304_305insGG (p.Ala102GlyfsTer28, rs201240957) in the HLA-DRB5 gene that was present as homozygous (n = 1) and heterozygous (n = 2) in inhibitor-positive patients. Another variant that was identified as homozygous (n = 1) and heterozygous (n = 2) in inhibitor-positive patients was the rs200975376 (c.134G>A, p.Arg45His) missense variant in the chemokine (C-C motif) ligand 4-like 1 (CCL4L1) gene. We further checked the presence of these mutations in 2 discordant brother-pairs present in our discovery phase, and we confirmed the rs200975376 variant in the CCL4L1 gene as a potential risk factor in the E835-E1309 brother-pair (supplemental Figure 1).
Rare (MAF ≤1%), damaging variants identified in immunoregulatory genes by WES of 26 Italian severe HA patients
| Gene . | Chr. . | Genomic position . | dbSNP ID . | Reference . | Alternate sequence . | Result . | Nucleotide change . | Amino acid change . | EUR MAF . | EA MAF . | Cases . | Controls . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk | ||||||||||||
| CD101 | 1 | 117552712 | rs371892797 | A | G | Missense | c.284A>G | p.Tyr95Cys | — | — | 1 het | — |
| IGKC | 2 | 89156939 | rs200765148 | A | C | Missense | c.258T>G | p.Cys87Gly | — | 0.001 | 1 het | — |
| IGKV1-39 | 2 | 89619806 | rs114472229 | G | A | Stop gained | c.52C>T | p.Arg18Ter | — | — | 3 het | — |
| CCR5 | 3 | 46414784 | rs34418657 | G | T | Missense | c.391G>T | p.Val131Phe | — | 0.0001 | 1 het | — |
| HLA-DRB5 | 6 | 32489747 | rs201240957 | — | GG | Frameshift | c.304_305insGG | p.Ala102GlyfsTer28 | — | — | 1 hom 2 het | — |
| HLA-DRB1 | 6 | 32551953 | rs9281873 | CG | — | Frameshift | c.301_302delCG | p.Arg101GlyfsTer26 | — | — | 1 het | — |
| 6 | 32726657 | rs202167169 | T | C | Missense | c.616A>G | p.Ser206Gly | — | — | 1 het | — | |
| CD83 | 6 | 14135348 | — | C | T | Missense | c.499C>T | p.Arg167Trp | — | — | 1 het | — |
| PMS2 | 7 | 6045633 | rs201343342 | A | G | Missense | c.53T>C | p.Ile18Thr | — | — | 2 het | — |
| CD36 | 7 | 80301310 | rs56381858 | T | G | Stop gained | c.1079T>G | p.Leu360Ter | — | 0.0001 | 1 het | — |
| NLRX1 | 11 | 119045378 | rs142087333 | C | T | Missense | c.1066C>T | p.Arg356Trp | 0.001 | 0.0004 | 2 het | — |
| IGHG3 | 14 | 106236141 | rs60746425 | G | A | Missense | c.663C>T | p.Arg222Trp | 0.001 | 0.003 | 1 het | — |
| 14 | 106237568 | rs201430154 | G | T | Missense | c.175C>A | p.Ser59Tyr | — | 0.002 | 1 het | — | |
| IGHG4 | 14 | 106090814 | rs376149342 | C | T | Stop retained | c.983G>A | c.983G>A | — | — | 1 het | — |
| IGHV3-53 | 14 | 107048725 | rs200210744 | T | — | Frameshift | c.294delT | p.Leu99PhefsTer3 | — | — | 1 het | — |
| IGHV4-4 | 14 | 106478358 | rs201063945 | A | G | Missense | c.100T>C | p.Ser34Pro | — | — | 1 het | — |
| CCL4L1 | 17 | 34539235 | rs200975376 | G | A | Missense | c.134G>A | p.Arg45His | — | — | 1 hom 1 het | — |
| Protective | ||||||||||||
| IGKV3-7 | 2 | 89278201 | rs376449387 | G | C | Missense | c.134C>G | p.Ala45Gly | — | — | — | 3 het |
| IGKV1-12 | 2 | 89339834 | rs201844946 | C | A | Missense | c.238G>T | p.Val80Phe | — | 0.001 | — | 1 het |
| CX3CR1 | 3 | 39307768 | — | — | C | Frameshift | c.328dupC | p.Leu110ProfsTer17 | — | — | — | 1 het |
| BST1 | 4 | 15724520 | rs144197373 | G | A | Missense | c.814G>A | p.Val272Met | 0.01 | 0.001 | — | 1 het |
| IL7R | 5 | 35876449 | rs2229232 | C | T | Missense | c.1241C>T | p.Thr414Met | 0.003 | 0.01 | — | 1 het |
| XRCC4 | 5 | 82649026 | rs61749611 | A | C | Missense | c.976A>C | p.Asn326His | — | 0.001 | — | 1 het |
| HLA-B | 6 | 31323184 | rs2308488 | C | T | Missense | c.805G>A | p.Ala269Thr | — | — | — | 1 het |
| CD109 | 6 | 74475822 | — | G | A | Missense | c.1277G>A | p.Gly426Glu | — | — | — | 1 het |
| IGHV4-28 | 14 | 106780667 | rs8010702 | T | G | Missense | c.197A>C | p.Glu66Ala | 0.01 | 0.002 | — | 1 het |
| IGHV3-38 | 14 | 106866451 | rs144366955 | G | C | Missense | c.303C>G | p.Asn101Lys | — | 0.003 | — | 1 het |
| IGHV6-1 | 14 | 106405706 | rs11849619 | G | A | Stop gained | c.268C>T | p.Arg90Ter | 0.003 | 0.004 | — | 1 het |
| Gene . | Chr. . | Genomic position . | dbSNP ID . | Reference . | Alternate sequence . | Result . | Nucleotide change . | Amino acid change . | EUR MAF . | EA MAF . | Cases . | Controls . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk | ||||||||||||
| CD101 | 1 | 117552712 | rs371892797 | A | G | Missense | c.284A>G | p.Tyr95Cys | — | — | 1 het | — |
| IGKC | 2 | 89156939 | rs200765148 | A | C | Missense | c.258T>G | p.Cys87Gly | — | 0.001 | 1 het | — |
| IGKV1-39 | 2 | 89619806 | rs114472229 | G | A | Stop gained | c.52C>T | p.Arg18Ter | — | — | 3 het | — |
| CCR5 | 3 | 46414784 | rs34418657 | G | T | Missense | c.391G>T | p.Val131Phe | — | 0.0001 | 1 het | — |
| HLA-DRB5 | 6 | 32489747 | rs201240957 | — | GG | Frameshift | c.304_305insGG | p.Ala102GlyfsTer28 | — | — | 1 hom 2 het | — |
| HLA-DRB1 | 6 | 32551953 | rs9281873 | CG | — | Frameshift | c.301_302delCG | p.Arg101GlyfsTer26 | — | — | 1 het | — |
| 6 | 32726657 | rs202167169 | T | C | Missense | c.616A>G | p.Ser206Gly | — | — | 1 het | — | |
| CD83 | 6 | 14135348 | — | C | T | Missense | c.499C>T | p.Arg167Trp | — | — | 1 het | — |
| PMS2 | 7 | 6045633 | rs201343342 | A | G | Missense | c.53T>C | p.Ile18Thr | — | — | 2 het | — |
| CD36 | 7 | 80301310 | rs56381858 | T | G | Stop gained | c.1079T>G | p.Leu360Ter | — | 0.0001 | 1 het | — |
| NLRX1 | 11 | 119045378 | rs142087333 | C | T | Missense | c.1066C>T | p.Arg356Trp | 0.001 | 0.0004 | 2 het | — |
| IGHG3 | 14 | 106236141 | rs60746425 | G | A | Missense | c.663C>T | p.Arg222Trp | 0.001 | 0.003 | 1 het | — |
| 14 | 106237568 | rs201430154 | G | T | Missense | c.175C>A | p.Ser59Tyr | — | 0.002 | 1 het | — | |
| IGHG4 | 14 | 106090814 | rs376149342 | C | T | Stop retained | c.983G>A | c.983G>A | — | — | 1 het | — |
| IGHV3-53 | 14 | 107048725 | rs200210744 | T | — | Frameshift | c.294delT | p.Leu99PhefsTer3 | — | — | 1 het | — |
| IGHV4-4 | 14 | 106478358 | rs201063945 | A | G | Missense | c.100T>C | p.Ser34Pro | — | — | 1 het | — |
| CCL4L1 | 17 | 34539235 | rs200975376 | G | A | Missense | c.134G>A | p.Arg45His | — | — | 1 hom 1 het | — |
| Protective | ||||||||||||
| IGKV3-7 | 2 | 89278201 | rs376449387 | G | C | Missense | c.134C>G | p.Ala45Gly | — | — | — | 3 het |
| IGKV1-12 | 2 | 89339834 | rs201844946 | C | A | Missense | c.238G>T | p.Val80Phe | — | 0.001 | — | 1 het |
| CX3CR1 | 3 | 39307768 | — | — | C | Frameshift | c.328dupC | p.Leu110ProfsTer17 | — | — | — | 1 het |
| BST1 | 4 | 15724520 | rs144197373 | G | A | Missense | c.814G>A | p.Val272Met | 0.01 | 0.001 | — | 1 het |
| IL7R | 5 | 35876449 | rs2229232 | C | T | Missense | c.1241C>T | p.Thr414Met | 0.003 | 0.01 | — | 1 het |
| XRCC4 | 5 | 82649026 | rs61749611 | A | C | Missense | c.976A>C | p.Asn326His | — | 0.001 | — | 1 het |
| HLA-B | 6 | 31323184 | rs2308488 | C | T | Missense | c.805G>A | p.Ala269Thr | — | — | — | 1 het |
| CD109 | 6 | 74475822 | — | G | A | Missense | c.1277G>A | p.Gly426Glu | — | — | — | 1 het |
| IGHV4-28 | 14 | 106780667 | rs8010702 | T | G | Missense | c.197A>C | p.Glu66Ala | 0.01 | 0.002 | — | 1 het |
| IGHV3-38 | 14 | 106866451 | rs144366955 | G | C | Missense | c.303C>G | p.Asn101Lys | — | 0.003 | — | 1 het |
| IGHV6-1 | 14 | 106405706 | rs11849619 | G | A | Stop gained | c.268C>T | p.Arg90Ter | 0.003 | 0.004 | — | 1 het |
Chr., chromosome; EA MAF, European American population from Exome Variant Server; EUR MAF, European population from the 1000 Genomes project; het, heterozygous; hom, homozygous.
Variants that increase or reduce the risk of inhibitor development
To identify other genetic factors associated with inhibitor development, a case-control association analysis was conducted between variants identified in inhibitor-positive patients (n = 17) and those identified in inhibitor-negative patients (n = 9) using different genetic models. To assess which genetic model best suited our data, we generated Q-Q plots and P value distribution histograms for each derived set of P values, and we found that the dominant and trend models produced the most normalized P value distributions (supplemental Figure 2). We then filtered out all the variants that did not reach statistical significance for both models (P > .05) and obtained a list of 1364 genetic variants potentially involved in inhibitor development.
To further analyze these 1364 genetic variants, we performed unsupervised hierarchical clustering that revealed 2 distinct patterns of homozygous variants (Figure 2): homozygous variants present in inhibitor-positive patients, indicating an elevated risk of inhibitor development (henceforth called risk), and 2 distinct clusters of homozygous variants present mainly in inhibitor-negative patients that can potentially have a protective role in inhibitor development (henceforth called protective). As shown in Figure 2, unsupervised hierarchical clustering correctly separated the majority of patients into 2 clusters: protective (black lines under the heatmap) and risk (red lines under the heatmap). Differences in modes of inheritance between the brothers are emphasized through pattern differences in the heatmap. Concordant brother-pairs E287-E1147 and E1361-E502 exhibited greater similarity across homo- and heterozygous variants and clustered with other inhibitor-negative and inhibitor-positive patients, respectively. On the other hand, the discordant brothers E835-E1309 and E172-E1355 exhibited greater differences across these variant types, which resulted in 2 brothers E835 and E1355 clustering away from the other risk and protective patients, respectively. This is most likely due to the close genetic background shared with their brothers, causing patient E835 to cluster together with his brother, E1309, in a protective cluster and E1355 together with E172 in a risk cluster (Figure 2). Biological relatedness between all these brother-pairs was confirmed based on all variants that passed QC using the Ajk statistic32 (supplemental Table 2B).
Clustering analysis (with heatmap) of variants identified in 26 Italian severe hemophilia A patients by WES. Unsupervised hierarchical clustering was performed using 1364 statistically significant variants (P ≤ .05) by Cochran-Armitage (trend model) and Fisher’s exact (dominant model) t tests. Two main, distinct patterns of homozygous variants were revealed that are characteristic of inhibitor-positive (risk) and inhibitor-negative (protective) patients. Differences in modes of inheritance between all brothers are emphasized through pattern differences in the heatmap: concordant brothers E287-E1147 (inhibitor negative; green rectangle) and E1361-E502 (inhibitor positive; orange rectangle) exhibit greater similarity across homo- and heterozygous variants, whereas the discordant brothers E835-E1309 and E172-E1355 (blue rectangles) exhibit greater differences across these variant types. Red bars under the heatmaps underline inhibitor-positive and black bars underline inhibitor-negative patients. Heatmap: red, homozygous variants; green, heterozygous variants; black, reference calls. Unsupervised hierarchical clustering was performed using R.31
Clustering analysis (with heatmap) of variants identified in 26 Italian severe hemophilia A patients by WES. Unsupervised hierarchical clustering was performed using 1364 statistically significant variants (P ≤ .05) by Cochran-Armitage (trend model) and Fisher’s exact (dominant model) t tests. Two main, distinct patterns of homozygous variants were revealed that are characteristic of inhibitor-positive (risk) and inhibitor-negative (protective) patients. Differences in modes of inheritance between all brothers are emphasized through pattern differences in the heatmap: concordant brothers E287-E1147 (inhibitor negative; green rectangle) and E1361-E502 (inhibitor positive; orange rectangle) exhibit greater similarity across homo- and heterozygous variants, whereas the discordant brothers E835-E1309 and E172-E1355 (blue rectangles) exhibit greater differences across these variant types. Red bars under the heatmaps underline inhibitor-positive and black bars underline inhibitor-negative patients. Heatmap: red, homozygous variants; green, heterozygous variants; black, reference calls. Unsupervised hierarchical clustering was performed using R.31
Identification of potentially damaging variants
Further filtering of these 1364 gene variants for damaging effects revealed a list of 80 variants, which we further trimmed by removing those that did not occur in canonical transcript38 or found in unknown genes such as annotated but unknown open reading frames or hypothetical proteins. This left us with 43 coding variants that were observed in 35 different genes. As shown in Table 2, most of the variants were common polymorphisms showing MAF >5%. Only 1 low-frequency variant rs2407221 (MAF, 3%) in the PRSS48 gene that encodes the STOP loss (Glu/Ter) has been identified. In addition, we found 2 rare variants, the rs6582584 in the ALG10B gene (MAF, 1%) and the rs148759495 in FCGBP gene, for which no MAF has been determined thus far. The affected pathways involve genes associated with the immune responses and autoimmunity, such as Immunoglobulin Heavy Constant γ 1 (IGHG1), Fc Fragment of IgG Binding Protein (FCGBP), Killer Cell Lectin-Like Receptor Subfamily B, Member 1 (KLRB1), and CD207 genes. To obtain a final list of variants for replication analysis, we excluded SNVs that were in high linkage disequilibrium (LD). Finally, we obtained a list of 28 nonsynonymous variants in 27 genes that were subjected to TaqMan SNP replication (Table 2). Variant filtering steps are graphically represented in Figure 1.
Nonsynonymous variants identified in a case-control association analysis using Cochran-Armitage and Fisher’s exact tests
| Gene . | Chr. . | Genomic position . | dbSNP ID . | TSI minor allele . | TSI MAF . | Result . | Protective effect . | OR (95% CI) . | MAF inhibitor . | MAF noninhibitor . | Variants replicated . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL11A1 | 1 | 103379918 | rs3753841 | G | 0.40 | Missense | P/L | 0.68 (0.21-2.2) | 0.35 | 0.44 | X |
| FNDC7 | 1 | 109268573 | rs4494160 | T | 0.31* | Missense | V/A | 0.1 (0.02-0.55) | 0.06 | 0.39 | X |
| FOXJ3 | 1 | 42693597 | rs343376 | A | 0.33* | Missense | V/A | 0.27 (0.07-1.04) | 0.15 | 0.39 | |
| CLCA1 | 1 | 86959173 | rs2791494 | T | 0.20 | Missense | M/T | 0.52 (0.14-1.9) | 0.21 | 0.33 | |
| PLA2R1 | 2 | 160808075 | rs3828323 | T | 0.42 | Missense | G/S | 1.8 (0.6-5.7) | 0.53 | 0.39 | X |
| CRYGB | 2 | 209007559 | rs796287 | T | 0.22 | Missense | I/L | 0.27 (0.07-1) | 0.15 | 0.39 | X |
| CD207 | 2 | 71058835 | rs741326 | G | 0.44 | Missense | V/A | 0.5 (0.2-1.6) | 0.38 | 0.56 | X |
| LCT | 2 | 136590746 | rs3754689 | T | 0.40 | Missense | V/I | 0.6 (0.2-1.9) | 0.32 | 0.44 | X |
| TRIM42 | 3 | 140407260 | rs9876490 | C | 0.47 | Missense | A/E | 0.52 (0.16-1.7) | 0.29 | 0.44 | X |
| TTLL3 | 3 | 9871030 | rs2290305 | T | 0.23 | Missense | M/R | 0.07 (0.02-0.28) | 0.12 | 0.67 | X |
| HHLA2 | 3 | 108072298 | rs6779254 | T | 0.28 | Missense | I/T | 0.65 (0.2-2.2) | 0.29 | 0.39 | X |
| FAM184B | 4 | 17660082 | rs1860596 | C | 0.36 | Missense | R/H | 3.2 (1-10.7) | 0.62 | 0.33 | X |
| TLR3 | 4 | 187004074 | rs3775291 | T | 0.31 | Missense | L/F | 0.7 (0.2-2.5) | 0.21 | 0.28 | X |
| PRSS48 | 4 | 152212603 | rs2407221 | T | 0.03 | Stop lost | */E | 0.1 (0.01-1) | 0.03 | 0.22 | X |
| KIF4B | 5 | 154395158 | rs6580126 | G | 0.22 | Missense | R/L | 0.45 (0.14-1.5) | 0.26 | 0.44 | X |
| CHSY3 | 5 | 129521126 | rs2015018 | A | 0.28 | Missense | D/G | 0.1 (0.02-0.55) | 0.06 | 0.39 | |
| RREB1 | 6 | 7211818 | rs1334576 | A | 0.26 | Missense | G/R | 0.38 (0.1-1.3) | 0.24 | 0.44 | X |
| BLACE | 7 | 155150472 | rs3922604 | G | 0.23* | Missense | R/W | 0.1 (0.01-1) | 0.03 | 0.22 | |
| PLEKHA1 | 10 | 124189197 | rs1045216 | A | 0.44 | Missense | T/A | 0.30 (0.09-1) | 0.32 | 0.61 | X |
| DCLRE1C | 10 | 14974905 | rs12768894 | C | 0.13 | Missense | H/R | 1 (0.3-3.2) | 0.32 | 0.33 | X |
| OR8K3 | 11 | 56086147 | rs960193 | T | 0.36 | Missense | L/R | 0.38 (0.12-1.24) | 0.32 | 0.56 | |
| OR8K1 | 11 | 56113516 | rs1905055 | T | 0.36 | Initiator codon variant | M/T | 0.38 (0.12-1.24) | 0.32 | 0.56 | |
| DNHD1 | 11 | 6549779 | rs4633449 | A | 0.41* | Missense | D/A | 0.44 (1.3-15) | 0.74 | 0.39 | X |
| DNHD1 | 11 | 6549995 | rs7480644 | A | 0.44 | Missense | N/S | 0.23 (0.07-0.72) | 0.26 | 0.61 | |
| DNHD1 | 11 | 6550004 | rs2344828 | A | 0.44 | Missense | Q/R | 0.23 (0.07-0.72) | 0.26 | 0.61 | |
| DNHD1 | 11 | 6550309 | rs2344829 | G | 0.44 | Missense | G/R | 0.23 (0.07-0.72) | 0.26 | 0.61 | X |
| MAP1LC3B2 | 12 | 117013730 | rs7303998 | G | 0.23* | 5′UTR | — | 0.26 (0.07-0.9) | 0.21 | 0.50 | X |
| KLRB1 | 12 | 9750669 | rs1135816 | G | 0.43 | Missense | I/T | 2.9 (0.9-10) | 0.53 | 0.28 | X |
| ALG10B | 12 | 38712142 | rs6582584 | C | 0.01* | Missense | A/G | 0.06 (0.003-1.3) | 0.02 | 0.19 | X |
| PSMD9 | 12 | 122326812 | rs2230681 | T | 0.13 | Missense | V/A | 0.06 (0.007-0.56) | 0.03 | 0.33 | X |
| APOBEC1 | 12 | 7805236 | rs2302515 | C | 0.13* | Missense | M/I | 0.1 (0.01-1) | 0.03 | 0.22 | X |
| SLCO1B7 | 12 | 21196424 | rs11045676 | A | 0.19* | Missense | F/Y | 0.32 (0.1-1.1) | 0.21 | 0.44 | |
| SLCO1B7 | 12 | 21207389 | rs11045699 | C | 0.19* | Missense | F/L | 0.32 (0.1-1.1) | 0.21 | 0.44 | X |
| OR10G3 | 14 | 22038659 | rs17792778 | T | 0.27 | Missense | S/G | 0.6 (0.17-1.9) | 0.27 | 0.39 | |
| STON2 | 14 | 81744736 | rs3813535 | A | 0.19 | Missense | S/P | 0.27 (0.06-1.1) | 0.12 | 0.33 | X |
| IGHG1 | 14 | 106209119 | rs1071803 | T | 0.32† | Missense | K/R | 0.27 (0.07-1) | 0.15 | 0.39 | |
| IGHG3 | 14 | 106236128 | rs12890621 | T | 0.31† | Missense | Y/F | 0.27 (0.07-1) | 0.15 | 0.39 | |
| ANKRD62 | 18 | 12115509 | rs4519391 | G | 0.3 | Missense | E/K | 0.34 (0.09-1.23) | 0.18 | 0.39 | X |
| ANKRD62 | 18 | 12125657 | rs7243248 | G | 0.35* | Missense | A/T | 0.41 (0.12-1.44) | 0.21 | 0.39 | |
| SLC14A2 | 18 | 43262359 | rs3745009 | A | 0.48 | Missense | A/T | 0.18 (0.05-0.65) | 0.32 | 0.72 | X |
| FCGBP | 19 | 40402285 | rs148759495 | A | — | Missense | P/L | 0.52 (0.2-1.7) | 0.29 | 0.44 | |
| FCGBP | 19 | 40405997 | rs7249743 | C | 0.12* | Missense | M/V | 0.52 (0.2-1.7) | 0.29 | 0.44 | |
| GTSE1 | 22 | 46722400 | rs140054 | T | 0.08* | Missense | W/R | 0.06 (0.004-1.6) | 0.02 | 0.19 | X |
| Gene . | Chr. . | Genomic position . | dbSNP ID . | TSI minor allele . | TSI MAF . | Result . | Protective effect . | OR (95% CI) . | MAF inhibitor . | MAF noninhibitor . | Variants replicated . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL11A1 | 1 | 103379918 | rs3753841 | G | 0.40 | Missense | P/L | 0.68 (0.21-2.2) | 0.35 | 0.44 | X |
| FNDC7 | 1 | 109268573 | rs4494160 | T | 0.31* | Missense | V/A | 0.1 (0.02-0.55) | 0.06 | 0.39 | X |
| FOXJ3 | 1 | 42693597 | rs343376 | A | 0.33* | Missense | V/A | 0.27 (0.07-1.04) | 0.15 | 0.39 | |
| CLCA1 | 1 | 86959173 | rs2791494 | T | 0.20 | Missense | M/T | 0.52 (0.14-1.9) | 0.21 | 0.33 | |
| PLA2R1 | 2 | 160808075 | rs3828323 | T | 0.42 | Missense | G/S | 1.8 (0.6-5.7) | 0.53 | 0.39 | X |
| CRYGB | 2 | 209007559 | rs796287 | T | 0.22 | Missense | I/L | 0.27 (0.07-1) | 0.15 | 0.39 | X |
| CD207 | 2 | 71058835 | rs741326 | G | 0.44 | Missense | V/A | 0.5 (0.2-1.6) | 0.38 | 0.56 | X |
| LCT | 2 | 136590746 | rs3754689 | T | 0.40 | Missense | V/I | 0.6 (0.2-1.9) | 0.32 | 0.44 | X |
| TRIM42 | 3 | 140407260 | rs9876490 | C | 0.47 | Missense | A/E | 0.52 (0.16-1.7) | 0.29 | 0.44 | X |
| TTLL3 | 3 | 9871030 | rs2290305 | T | 0.23 | Missense | M/R | 0.07 (0.02-0.28) | 0.12 | 0.67 | X |
| HHLA2 | 3 | 108072298 | rs6779254 | T | 0.28 | Missense | I/T | 0.65 (0.2-2.2) | 0.29 | 0.39 | X |
| FAM184B | 4 | 17660082 | rs1860596 | C | 0.36 | Missense | R/H | 3.2 (1-10.7) | 0.62 | 0.33 | X |
| TLR3 | 4 | 187004074 | rs3775291 | T | 0.31 | Missense | L/F | 0.7 (0.2-2.5) | 0.21 | 0.28 | X |
| PRSS48 | 4 | 152212603 | rs2407221 | T | 0.03 | Stop lost | */E | 0.1 (0.01-1) | 0.03 | 0.22 | X |
| KIF4B | 5 | 154395158 | rs6580126 | G | 0.22 | Missense | R/L | 0.45 (0.14-1.5) | 0.26 | 0.44 | X |
| CHSY3 | 5 | 129521126 | rs2015018 | A | 0.28 | Missense | D/G | 0.1 (0.02-0.55) | 0.06 | 0.39 | |
| RREB1 | 6 | 7211818 | rs1334576 | A | 0.26 | Missense | G/R | 0.38 (0.1-1.3) | 0.24 | 0.44 | X |
| BLACE | 7 | 155150472 | rs3922604 | G | 0.23* | Missense | R/W | 0.1 (0.01-1) | 0.03 | 0.22 | |
| PLEKHA1 | 10 | 124189197 | rs1045216 | A | 0.44 | Missense | T/A | 0.30 (0.09-1) | 0.32 | 0.61 | X |
| DCLRE1C | 10 | 14974905 | rs12768894 | C | 0.13 | Missense | H/R | 1 (0.3-3.2) | 0.32 | 0.33 | X |
| OR8K3 | 11 | 56086147 | rs960193 | T | 0.36 | Missense | L/R | 0.38 (0.12-1.24) | 0.32 | 0.56 | |
| OR8K1 | 11 | 56113516 | rs1905055 | T | 0.36 | Initiator codon variant | M/T | 0.38 (0.12-1.24) | 0.32 | 0.56 | |
| DNHD1 | 11 | 6549779 | rs4633449 | A | 0.41* | Missense | D/A | 0.44 (1.3-15) | 0.74 | 0.39 | X |
| DNHD1 | 11 | 6549995 | rs7480644 | A | 0.44 | Missense | N/S | 0.23 (0.07-0.72) | 0.26 | 0.61 | |
| DNHD1 | 11 | 6550004 | rs2344828 | A | 0.44 | Missense | Q/R | 0.23 (0.07-0.72) | 0.26 | 0.61 | |
| DNHD1 | 11 | 6550309 | rs2344829 | G | 0.44 | Missense | G/R | 0.23 (0.07-0.72) | 0.26 | 0.61 | X |
| MAP1LC3B2 | 12 | 117013730 | rs7303998 | G | 0.23* | 5′UTR | — | 0.26 (0.07-0.9) | 0.21 | 0.50 | X |
| KLRB1 | 12 | 9750669 | rs1135816 | G | 0.43 | Missense | I/T | 2.9 (0.9-10) | 0.53 | 0.28 | X |
| ALG10B | 12 | 38712142 | rs6582584 | C | 0.01* | Missense | A/G | 0.06 (0.003-1.3) | 0.02 | 0.19 | X |
| PSMD9 | 12 | 122326812 | rs2230681 | T | 0.13 | Missense | V/A | 0.06 (0.007-0.56) | 0.03 | 0.33 | X |
| APOBEC1 | 12 | 7805236 | rs2302515 | C | 0.13* | Missense | M/I | 0.1 (0.01-1) | 0.03 | 0.22 | X |
| SLCO1B7 | 12 | 21196424 | rs11045676 | A | 0.19* | Missense | F/Y | 0.32 (0.1-1.1) | 0.21 | 0.44 | |
| SLCO1B7 | 12 | 21207389 | rs11045699 | C | 0.19* | Missense | F/L | 0.32 (0.1-1.1) | 0.21 | 0.44 | X |
| OR10G3 | 14 | 22038659 | rs17792778 | T | 0.27 | Missense | S/G | 0.6 (0.17-1.9) | 0.27 | 0.39 | |
| STON2 | 14 | 81744736 | rs3813535 | A | 0.19 | Missense | S/P | 0.27 (0.06-1.1) | 0.12 | 0.33 | X |
| IGHG1 | 14 | 106209119 | rs1071803 | T | 0.32† | Missense | K/R | 0.27 (0.07-1) | 0.15 | 0.39 | |
| IGHG3 | 14 | 106236128 | rs12890621 | T | 0.31† | Missense | Y/F | 0.27 (0.07-1) | 0.15 | 0.39 | |
| ANKRD62 | 18 | 12115509 | rs4519391 | G | 0.3 | Missense | E/K | 0.34 (0.09-1.23) | 0.18 | 0.39 | X |
| ANKRD62 | 18 | 12125657 | rs7243248 | G | 0.35* | Missense | A/T | 0.41 (0.12-1.44) | 0.21 | 0.39 | |
| SLC14A2 | 18 | 43262359 | rs3745009 | A | 0.48 | Missense | A/T | 0.18 (0.05-0.65) | 0.32 | 0.72 | X |
| FCGBP | 19 | 40402285 | rs148759495 | A | — | Missense | P/L | 0.52 (0.2-1.7) | 0.29 | 0.44 | |
| FCGBP | 19 | 40405997 | rs7249743 | C | 0.12* | Missense | M/V | 0.52 (0.2-1.7) | 0.29 | 0.44 | |
| GTSE1 | 22 | 46722400 | rs140054 | T | 0.08* | Missense | W/R | 0.06 (0.004-1.6) | 0.02 | 0.19 | X |
Variants in high LD are in bold.
Chr., chromosome; UTR, untranslated region.
MAF from HapMap-CEU of Utah residence with Northern and Western European ancestry form HapMap phase 3 project given when TSI is not present.
MAF from CSAgilent population of European descent from the ClinSeq project given when neither HapMap TSI or CEU are present.
Replication of the 28 nonsynonymous variants identified in WES
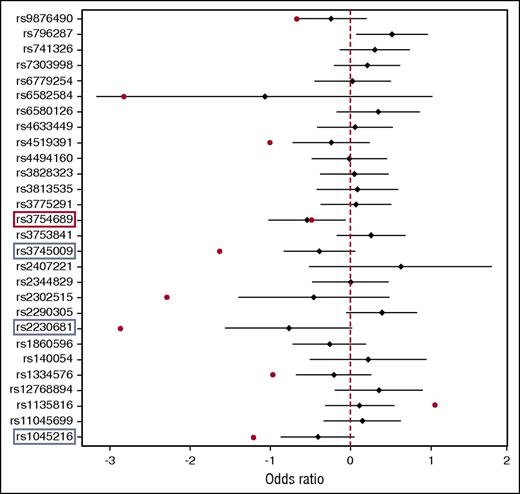
To validate the WES data, we replicated the 28 SNVs in 53 inhibitor-positive and 174 inhibitor-negative severe HA patients, and we performed logistic regression association analysis. As shown in Figure 3, we identified 10 variants that showed the same trend with the WES study. The remaining 18 variants were either equally distributed between inhibitor-positive and inhibitor-negative patients or were discordant with the sequencing results. The top variant replicated was rs3754689 in the LCT gene, which was significantly associated with inhibitor development (OR, 0.58; 95% CI, 0.36-0.94; P = .028). The rs3754689 SNV encodes C>T missense change that results in a valine to isoleucine substitution at position p.Val219Ile of the lactase protein. We found the rs3754689-T allele in 27 inhibitor-positive and 106 inhibitor-negative patients, corresponding to a MAF of 0.26 and 0.38 for inhibitor-positive and inhibitor-negative patients, respectively. This result confirmed the protective role for the rs3754689-T allele previously seen in WES with an OR of 0.6 (Table 3). However, adjusting for multiple testing using Benjamini and Hochberg’s method resulted in a false discovery rate of 22%. We also performed a separate analysis for all unrelated patients from the replication cohort (n = 185), of whom 47 were inhibitor positive and 138 were inhibitor negative. The results confirmed once again the protective effect of the rs3754689-T allele in inhibitor development (OR, 0.56; 95% CI, 0.33-0.94; P = .028). Similar results were obtained from the association analysis of 41 HR patients and 174 inhibitor-negative patients (OR, 0.56; 95% CI, 0.33-0.95; P = .032).
Forest plot association analysis of the 28 nonsynonymous variants identified in WES and replicated in 227 severe HA patients. TaqMan SNP genotyping-based replication was performed in 53 inhibitor-positive and 174 inhibitor-negative patients. Data analysis was performed using logistic regression analysis adjusting for first-degree relatives. Black diamonds denote OR estimates, and the black bars denote the range of 95% CIs of each variant replicated. Red dots denote OR of variants showing the same trend with WES. Red rectangle denotes statistically significant association, and blue rectangles denote variants with borderline P values. The graph was generated using R.31
Forest plot association analysis of the 28 nonsynonymous variants identified in WES and replicated in 227 severe HA patients. TaqMan SNP genotyping-based replication was performed in 53 inhibitor-positive and 174 inhibitor-negative patients. Data analysis was performed using logistic regression analysis adjusting for first-degree relatives. Black diamonds denote OR estimates, and the black bars denote the range of 95% CIs of each variant replicated. Red dots denote OR of variants showing the same trend with WES. Red rectangle denotes statistically significant association, and blue rectangles denote variants with borderline P values. The graph was generated using R.31
Nonsynonymous variants replicated in 227 severe hemophilia A patients and showing a direction of association consistent with discovery phase
| Gene . | Chr. . | dbSNP ID . | Minor allele . | Result . | Protein effect . | WES . | Replication . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF inhibitor . | MAF noninhibitor . | OR (95% CI) . | MAF inhibitor . | MAF noninhibitor . | OR (95% CI) . | P value* (all patients) . | P value† (unrelated patients) . | ||||||
| LCT | 2 | rs3754689 | T | Missense | V/I | 0.32 | 0.44 | 0.60 (0.2-1.9) | 0.26 | 0.38 | 0.58 (0.36-0.94) | .028 | .028 |
| TRIM42 | 3 | rs9876490 | C | Missense | A/E | 0.29 | 0.44 | 0.52 (0.16-1.7) | 0.38 | 0.44 | 0.79 (0.51-1.23) | .30 | .13 |
| RREB1 | 6 | rs1334576 | A | Missense | G/R | 0.24 | 0.44 | 0.38 (0.1-1.3) | 0.29 | 0.33 | 0.81 (0.51-1.3) | .38 | .19 |
| PLEKHA1 | 10 | rs1045216 | A | Missense | T/A | 0.32 | 0.61 | 0.30 (0.09-1) | 0.31 | 0.40 | 0.67 (0.42-1.1) | .081 | .12 |
| ALG10B | 12 | rs6582584 | C | Missense | A/G | 0.02 | 0.19 | 0.06 (0.003-1.3) | 0.01 | 0.03 | 0.34 (0.04-2.8) | .32 | .50 |
| APOBEC1 | 12 | rs2302515 | C | Missense | M/I | 0.03 | 0.22 | 0.10 (0.01-1) | 0.05 | 0.07 | 0.63 (0.25-1.6) | .34 | .49 |
| KLRB1 | 12 | rs1135816 | G | Missense | I/T | 0.53 | 0.28 | 2.90 (0.9-10) | 0.46 | 0.43 | 1.12 (0.73-1.7) | .60 | .60 |
| PSMD9 | 12 | rs2230681 | T | Missense | V/A | 0.03 | 0.33 | 0.06 (0.01-0.56) | 0.08 | 0.15 | 0.47 (0.21-1.02) | .056 | .14 |
| ANKRD62 | 18 | rs4519391 | G | Missense | E/K | 0.18 | 0.39 | 0.34 (0.09-1.23) | 0.30 | 0.35 | 0.78 (0.49-1.27) | .33 | .18 |
| SLC14A2 | 18 | rs3745009 | A | Missense | A/T | 0.32 | 0.72 | 0.18 (0.05-0.65) | 0.38 | 0.47 | 0.68 (0.44-1.06) | .087 | .047 |
| Gene . | Chr. . | dbSNP ID . | Minor allele . | Result . | Protein effect . | WES . | Replication . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF inhibitor . | MAF noninhibitor . | OR (95% CI) . | MAF inhibitor . | MAF noninhibitor . | OR (95% CI) . | P value* (all patients) . | P value† (unrelated patients) . | ||||||
| LCT | 2 | rs3754689 | T | Missense | V/I | 0.32 | 0.44 | 0.60 (0.2-1.9) | 0.26 | 0.38 | 0.58 (0.36-0.94) | .028 | .028 |
| TRIM42 | 3 | rs9876490 | C | Missense | A/E | 0.29 | 0.44 | 0.52 (0.16-1.7) | 0.38 | 0.44 | 0.79 (0.51-1.23) | .30 | .13 |
| RREB1 | 6 | rs1334576 | A | Missense | G/R | 0.24 | 0.44 | 0.38 (0.1-1.3) | 0.29 | 0.33 | 0.81 (0.51-1.3) | .38 | .19 |
| PLEKHA1 | 10 | rs1045216 | A | Missense | T/A | 0.32 | 0.61 | 0.30 (0.09-1) | 0.31 | 0.40 | 0.67 (0.42-1.1) | .081 | .12 |
| ALG10B | 12 | rs6582584 | C | Missense | A/G | 0.02 | 0.19 | 0.06 (0.003-1.3) | 0.01 | 0.03 | 0.34 (0.04-2.8) | .32 | .50 |
| APOBEC1 | 12 | rs2302515 | C | Missense | M/I | 0.03 | 0.22 | 0.10 (0.01-1) | 0.05 | 0.07 | 0.63 (0.25-1.6) | .34 | .49 |
| KLRB1 | 12 | rs1135816 | G | Missense | I/T | 0.53 | 0.28 | 2.90 (0.9-10) | 0.46 | 0.43 | 1.12 (0.73-1.7) | .60 | .60 |
| PSMD9 | 12 | rs2230681 | T | Missense | V/A | 0.03 | 0.33 | 0.06 (0.01-0.56) | 0.08 | 0.15 | 0.47 (0.21-1.02) | .056 | .14 |
| ANKRD62 | 18 | rs4519391 | G | Missense | E/K | 0.18 | 0.39 | 0.34 (0.09-1.23) | 0.30 | 0.35 | 0.78 (0.49-1.27) | .33 | .18 |
| SLC14A2 | 18 | rs3745009 | A | Missense | A/T | 0.32 | 0.72 | 0.18 (0.05-0.65) | 0.38 | 0.47 | 0.68 (0.44-1.06) | .087 | .047 |
Logistic regression analysis adjusting for first-degree relatives was used in replication.
Chr., chromosome.
P value from all 227 patients (n = 53 with and n = 174 without inhibitors).
P value from 185 unrelated patients (n = 47 with and n = 138 without inhibitors).
In addition, logistic regression analysis revealed 3 missense variants with borderline P values: rs1045216 in the PLEKHA1 gene (OR, 0.67; 95% CI, 0.42-1.1; P = .08), rs2230681 in the PSMD9 gene (OR, 0.47; 95% CI, 0.21-1.02; P = .056), and rs3745009 in the SLC14A2 gene (OR, 0.68; 95% CI, 0.44-1.06; P = .087). All 3 variants showed effect direction concordant with those previously seen in WES (Table 3).
Discussion
In this article, we report the results of WES and subsequent downstream data analysis to identify novel genetic risk factors for inhibitor development in 26 Italian patients with severe HA. We identified several rare, damaging variants present in immunoregulatory genes, as well as 28 nonsynonymous variants with putative roles in inhibitor development obtained from a case-control association analysis. Replication of these 28 variants in a larger cohort of 227 severe HA patients confirmed the protective effect of the rs3754689 missense variant in the LCT gene (OR, 0.58; 95% CI, 0.36-0.94; P = .028) as putative susceptibility locus for inhibitor development.
The development of neutralizing antibodies toward FVIII involves several steps of immune activation that include endocytosis and endosomal processing of FVIII by antigen presenting cells like dendritic or B cells, presentation of these peptides via the HLA class II molecules to the CD4+ T cells, and T cell–dependent stimulation of B-cell differentiation into antibody-secreting plasma or memory cells.39 The genetic variants identified in WES could potentially disrupt immune activation at any of these steps, leading to either increased or decreased risk of developing FVIII alloantibodies. For instance, we found variants in the HLA class II genes, HLA DRB1 and HLA DRB5, which have an essential role in presenting FVIII peptides to CD4+ T-helper cells.11-14 The HLA DRB1 locus is particularly interesting as it has been shown to be, together with the HLA DQB1, the most consistent risk factor in inhibitor development.9,13 We also identified variants that may affect the production or function of the immunoglobulin molecules: 5 immunoglobulin heavy chain variable regions, 3 variants in the immunoglobulin κ variable cluster, and SNVs in immunoglobulin constant heavy γ chains (Table 1). Indeed, previous studies demonstrated that class switch recombination is necessary to produce IgG1 and IgG4, the main subclasses of FVIII neutralizing antibodies, by activated B cells.40,41 Moreover, we found variants in genes encoding DNA repair molecules like XRCC4 and PMS2. The Xrcc4 protein is involved in nonhomologous end joining, a pathway used to repair double-stranded DNA breaks generated during V(D)J recombination that occurs between the immunoglobulin and T-cell receptor variable regions in developing lymphocytes.42 Other variants identified by WES may affect cytokines, ligand/receptor chemokine subunits, interleukins, and other immune system modulators that are involved in various steps of immune system activation and inflammation.
The LCT gene encodes the lactase-phlorizin hydrolase, an enzyme with both lactase and phlorizin hydrolase activity,43,44 which is required for the digestion of lactose sugar in the small intestine of humans and other mammals. The rs3754689 missense variant leads to a valine to isoleucine amino acid substitution at position p.Val219Ile of the lactase protein, and it has been predicted by both SIFT and PolyPhen-2 as benign. Indeed, this amino acid change has been previously reported by Boll at al44 as a polymorphism.
If rs3754689 is not causal, it may be coinherited with other causal SNV(s) that affect genes involved in immune responses. To answer this question, we carried out LD analysis of all 28 nonsynonymous SNVs in our replication study, as well as WES data, and found no variants in immunoregulatory genes that were coinherited with this SNV (data not shown). However, when the LD analysis was expanded to include variants in the genomic region surrounding the main association on chromosome 2q21, we found 9 SNVs present in the same individuals as the rs3754689 variant, indicating that they are in high LD with each other. In addition to the 2 intron variants in the LCT gene, we found 5 variants located in 2 downstream genes, R3HDM1 and UBXN4, as well as 2 intron variants located in 2 upstream genes: MCM6 and DARS. Such high LD in this region is not surprising because the haplotype surrounding the LCT locus underwent strong selective pressure in certain populations as a result of establishment of lactase persistence, a heritable genetic trait in which intestinal lactase production persists into the adulthood.45 In the European populations, lactase persistence is largely attributed to the polymorphism in intron 13 of the MCM6 gene (C>T at position −13910 to the LCT gene; rs4988235) located in the cis-acting enhancer element that influences transcriptional activity of the neighboring LCT promoter.46,47 This implies that the association signal coming from the LCT locus may indicate adjacent loci within the same haplotype. Indeed, the R3HDM1 locus has been recently identified in pan-meta-genome-wide association studies as a putative susceptibility locus for 2 autoimmune disorders: systemic sclerosis and systemic lupus erythematosus.48 The Ubxn4 protein has been suggested to function in endoplasmic reticulum–associated protein degradation,49 a pathway that can potentially be involved in presenting the FVIII peptides to CD4+ T-helper cells by antigen-presenting cells.
Previous studies have identified several loci, such as IL10 (−1082 G>A), CTLA4 (−318 C>T), TNFA (−308 G>A), and others that are located in intergenic, upstream promoter regions or intronic sequences of immunoregulatory genes, as genetic susceptible loci for inhibitor development.9 Because our analysis was focused on the coding areas of the genome, most of these variants were not detected. We searched for previously identified coding variants in the immune genes and found rs231775 (CTLA4),50 rs2476601 (PTPN22),50 rs2879097 (PCGF2),51 rs1882019 (HSP90B1),51 and p.R131H SNV in the FCGR2 gene.20 None of these SNVs, however, were associated with inhibitor development in our discovery phase (data not shown), and the most plausible explanation is a lack of statistical power due to small discovery population.
Another limitation of our study is the wide range of CIs observed in the replication phase (Figure 2), which may imply small replication cohort and/or population stratification. Our replication population consisted of 227 severe HA patients, and according to our post hoc data analysis, this cohort size should have been sufficient to replicate the 28 variants identified in WES with nearly 100% power at 0.05 significance. However, one needs to bear in mind that, due to limited and unbalanced size, the MAF obtained from our discovery phase might have been imprecise, thereby affecting the replication population power analysis. To reduce potential confounding due to population stratification, we included in our replication cohort mainly Italian patients. Only 6 patients (2.6%) of the entire replication cohort were of geographic origin other than Italian. Such a low ethnic diversity could not have had profound effects on data analysis. In fact, logistic regression association analysis excluding patients with non-Italian origin confirmed once again the rs3754689 missense variant (OR, 0.55; 95% CI, 0.34-0.91; P = .019).
In conclusion, we report the identification of several rare, damaging variants in immunoregulatory molecules, as well as the rs3754689 missense variant in the LCT gene, as putative susceptibility locus for inhibitor development. Given the extensive linkage surrounding the LCT locus due to a conserved haplotype, the association signal captured by the rs3754689 variant may belong to the adjacent loci such as the R3HDM1 or UBXN4. Further research is needed to pinpoint the exact signal coming from this conserved haplotype on chromosome 2q21 and to unravel the immunological mechanisms underlying the association between this variant(s) and risk of inhibitor development. Our findings might help to define the genetic landscape of inhibitor risk and promote the development of genetic risk algorithms, which will take into account all known genetic risk factors. The rapid advent of innovation technologies, such as exome/genome sequencing, will enable the identification of yet unknown genetic risk factors and pathways involved in inhibitor development. Use of such algorithms may allow to stratify patients for risk of inhibitor development that will permit to advance personalized treatment and clinical management strategies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the following colleagues from the Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Fondazione IRCCS Ca’ Granda Ospedale Maggiore: A. Cairo and S. Seregni for help with genetic analysis and M. Boscarino for help in the preparation of the figures; M. Fornili and E. M. Biganzoli (Dipartimento di Scienze Cliniche e di Comunità, University of Milan, Milan, Italy) for help with statistical analysis; and F. R. Rosendaal (Departments of Clinical Epidemiology, Thrombosis and Hemostasis, Leiden University Medical Center, Leiden, The Netherlands) for critical revision of the manuscript and its methodology.
This research project has been founded by a Bayer Hemophilia Special Project Award 2011 entitled “Discovery of genetic determinants of inhibitor development in hemophilia A by exome sequencing” awarded to Flora Peyvandi.
Authorship
Contribution: M.M.G. and L.A.L. designed the research and, together with K.B., performed data analysis, interpreted the results, and drafted the manuscript; E.P. performed experiments and, together with I.G., collected data and helped with the interpretation of the results; I.M. helped with data analysis; M.R.F., M.E.M., and E.S. were involved in recruiting patients; and F.P. designed the project, supervised the research, interpreted the results, and critically reviewed the manuscript.
Conflict-of-interest disclosure: F.P. has received honoraria for participating as a speaker at satellite symposia and educational meetings organized by Bayer, Biotest, CSL Behring, Grifols, Novo Nordisk, and Sobi. She is the recipient of research grant funding from Alexion, Biotest, Kedrion Biopharma, and Novo Nordisk paid to Fondazione Luigi Villa, and she has received consulting fees from Kedrion Biopharma, LFB, and Octapharma. She is a member of the Ablynx scientific advisory board. E.S. received fees as a speaker in meetings organized by Kedrion; acted as a paid consultant for Bayer, Pfizer, CSL Behring, NovoNordisk, Grifols, Baxter, Baxalta, Biogen Idec, Sobi, Octapharma, and Roche; and received an unrestricted research grant from Pfizer. M.E.M. received fees as a speaker in meetings organized by Bayer, CSL Behring, Novo Nordisk, Pfizer, and Sobi/Biogen Idec and acted as a paid consultant for Bayer, Pfizer, CSL Behring, Novo Nordisk, Baxter/Baxalta, and Sobi/Biogen Idec. The remaining authors declare no competing financial interests.
Correspondence: Flora Peyvandi, Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Fondazione IRCCS Ca' Granda, Ospedale Maggiore Policlinico and Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Via Pace 9, 20122 Milan, Italy; e-mail: flora.peyvandi@unimi.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal