Abstract
Gene editing is a rapidly developing area of biotechnology in which the nucleotide sequence of the genome of living cells is precisely changed. The use of genome-editing technologies to modify various types of blood cells, including hematopoietic stem cells, has emerged as an important field of therapeutic development for hematopoietic disease. Although these technologies offer the potential for generation of transformative therapies for patients suffering from myriad disorders of hematopoiesis, their application for therapeutic modification of primary human cells is still in its infancy. Consequently, development of ethical and regulatory frameworks that ensure their safe and effective use is an increasingly important consideration. Here, we review a number of issues that have the potential to impact the clinical implementation of genome-editing technologies, and suggest paths forward for resolving them such that new therapies can be safely and rapidly translated to the clinic.
Introduction
The ability to specifically modify individual loci in the genomes of patients' cells (gene editing) provides an ideal method to correct inherited disorders. Although gene editing can be done using adeno-associated virus (AAV) vectors or even synthetic nucleotide templates,1 for the purposes of this review, we will focus on gene editing that is mediated through an engineered nuclease. In nuclease-mediated gene editing, an engineered nuclease, from any nuclease platform technology (Table 1), is used to create a specific double-strand break (DSB) in the genome of a cell.2,3 This DSB activates the cells’ DNA repair machinery (Figure 1), and if the break is repaired in a mutagenic fashion by nonhomologous end joining (NHEJ), insertions/deletions can be generated at a specific genomic location, thereby giving precise spatial resolution to the modification. If, on the other hand, the DSB is repaired by homologous recombination (HR; also called homology-directed repair [HDR]) using a provided donor template that can enter into the HR process and input new genetic information into the target genome, specific nucleotide changes can be made to the genome at a specific genomic location thereby giving precise spatial and nucleotide resolution to the modification. In HR-mediated genome editing, single nucleotides can be changed or whole multigene cassettes can be inserted into the genome in a precise location.
Engineered nuclease platform technologies
| Nuclease . | Properties . | Advantages . | Disadvantages . |
|---|---|---|---|
| ZFN | Heterodimeric protein with multimeric sequence-specific zinc finger–binding domains and the Fok1 endonuclease | Most clinically advanced platform. | De novo design of effective and specific ZFNs is difficult |
| Can iteratively improve activity and specificity | |||
| Homing endonuclease (HE) | Homo- or dimeric protein containing site- specific DNA recognition domains and endonucleolytic activity | Smallest coding sequence; compatible with all vector platforms | Have been challenging to direct to new specific target sequences |
| High activity and specificity from complex protein structure | |||
| Unique 3′ overhang may have unique in vivo biochemistry | |||
| TALEN | Heterodimeric protein with multimeric site-specific TAL effector domains and the Fok1 endonuclease | Relatively easy to design and produce | Variable activity and specificity. May be able to evolve in vitro to higher efficacy; repetitive structure and large size render vectorization more complex |
| CRISPR | Combines a protein (with both RNA- directed DNA sequence-specific binding and endonuclease activity) and a short RNA guide sequence that recognizes the target site and directs the DNA binding of the nuclease | Easy to design to target new sequences; use as ribonucleoprotein particle may allow efficient modification of cells where translation rates are low | Need further knowledge of rare off-target events, frequency, and potential genotoxic effects; need for 2 biochemically different components complicates vectorization for some applications |
| Mega-TAL | Monomeric protein with TAL effector DNA-binding domain fused to a homing (mega-) endonuclease cleavage domain | Monomeric, compact albeit larger than HE alone, high activity and specificity from HE domain | Requires engineering a HE cleavage domain |
| Nuclease . | Properties . | Advantages . | Disadvantages . |
|---|---|---|---|
| ZFN | Heterodimeric protein with multimeric sequence-specific zinc finger–binding domains and the Fok1 endonuclease | Most clinically advanced platform. | De novo design of effective and specific ZFNs is difficult |
| Can iteratively improve activity and specificity | |||
| Homing endonuclease (HE) | Homo- or dimeric protein containing site- specific DNA recognition domains and endonucleolytic activity | Smallest coding sequence; compatible with all vector platforms | Have been challenging to direct to new specific target sequences |
| High activity and specificity from complex protein structure | |||
| Unique 3′ overhang may have unique in vivo biochemistry | |||
| TALEN | Heterodimeric protein with multimeric site-specific TAL effector domains and the Fok1 endonuclease | Relatively easy to design and produce | Variable activity and specificity. May be able to evolve in vitro to higher efficacy; repetitive structure and large size render vectorization more complex |
| CRISPR | Combines a protein (with both RNA- directed DNA sequence-specific binding and endonuclease activity) and a short RNA guide sequence that recognizes the target site and directs the DNA binding of the nuclease | Easy to design to target new sequences; use as ribonucleoprotein particle may allow efficient modification of cells where translation rates are low | Need further knowledge of rare off-target events, frequency, and potential genotoxic effects; need for 2 biochemically different components complicates vectorization for some applications |
| Mega-TAL | Monomeric protein with TAL effector DNA-binding domain fused to a homing (mega-) endonuclease cleavage domain | Monomeric, compact albeit larger than HE alone, high activity and specificity from HE domain | Requires engineering a HE cleavage domain |
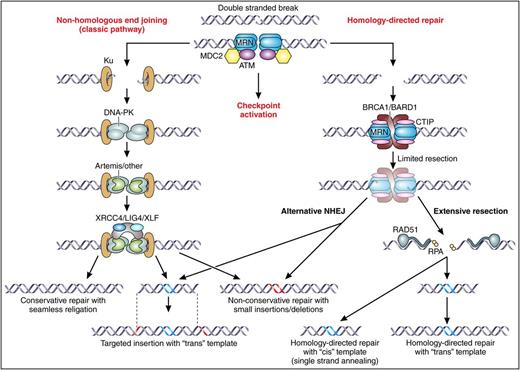
DNA DSB repair. The central initiating event in gene editing is a DNA DSB, which may be resolved through 2 different branches of cellular DNA repair mechanisms. The first branch is known as classic NHEJ, which primarily acts to directly rejoin the broken ends. This may occur seamlessly, with the generation of small insertions or deletions at the ends of the break (red-colored DNA), or in the case that the gene-editing procedure involves provision of a piece of foreign DNA, by insertion of a “trans” template into the DSB (blue-colored DNA in the pictured “trans” insertion template represents nonnative DNA sequence, gray DNA represents regions of homology to each side of the DSB; these would be optional if the “trans template” were intended solely for targeted insertion, but would be included in a typical gene targeting construct). The second branch is known as HDR, which involves the resection at the 5′ ends of each side of the DSB, and use of the resulting single-stranded DNA ends in a homology search for complementary DNA sequences. This may result in identification of short strands of homology near the original break site, which, upon resealing of the ends, will typically result in small deletions through a process known as alternative NHEJ (note that this pathway may also lead to targeted insertion of foreign DNA). Alternatively, extensive resection may occur, following which more extensive regions of homologous base pairing are required to resolve the break through HDR with a trans or cis template. Professional illustration by Patrick Lane, ScEYEnce Studios.
DNA DSB repair. The central initiating event in gene editing is a DNA DSB, which may be resolved through 2 different branches of cellular DNA repair mechanisms. The first branch is known as classic NHEJ, which primarily acts to directly rejoin the broken ends. This may occur seamlessly, with the generation of small insertions or deletions at the ends of the break (red-colored DNA), or in the case that the gene-editing procedure involves provision of a piece of foreign DNA, by insertion of a “trans” template into the DSB (blue-colored DNA in the pictured “trans” insertion template represents nonnative DNA sequence, gray DNA represents regions of homology to each side of the DSB; these would be optional if the “trans template” were intended solely for targeted insertion, but would be included in a typical gene targeting construct). The second branch is known as HDR, which involves the resection at the 5′ ends of each side of the DSB, and use of the resulting single-stranded DNA ends in a homology search for complementary DNA sequences. This may result in identification of short strands of homology near the original break site, which, upon resealing of the ends, will typically result in small deletions through a process known as alternative NHEJ (note that this pathway may also lead to targeted insertion of foreign DNA). Alternatively, extensive resection may occur, following which more extensive regions of homologous base pairing are required to resolve the break through HDR with a trans or cis template. Professional illustration by Patrick Lane, ScEYEnce Studios.
The use of genome-editing technologies to modify various types of blood cells, including hematopoietic stem cells (HSCs), has emerged as an important field of therapeutic development for hematopoietic disease.4,5 Although these technologies offer the potential for generation of transformative therapies for patients suffering from myriad disorders of hematopoiesis, their application for therapeutic modification of primary human cells is still in its infancy. Consequently, development of ethical and regulatory frameworks that ensure their safe and effective use is an increasingly important consideration. Here, we review a number of issues that have the potential to impact the clinical implementation of genome-editing technologies, and suggest paths forward for resolving them so that new therapies can be safely and rapidly translated to the clinic.
Ethical considerations in therapeutic gene editing
Although genome editing is a general process, the ethical implications of editing are not simply about the process, but instead are directly related to the purpose for which it is used. In “Germ line” and “Somatic cells,” we outline some thoughts on the ethical considerations of using genome editing to modify the germ line and somatic cells.
The ethical issues surrounding the use of gene modification that may affect the human germ line have been extensively discussed previously.6-11 The National Academy of Sciences (NAS) and National Academy of Medicine (NAM) in conjunction with the Royal Academy of Sciences and Chinese Academy of Sciences have assembled a “Committee on Human Gene Editing: Scientific, Medical, and Ethical Considerations” chaired by Professors Alta Charo and Richard Hynes to perform a year-long, in-depth study of the issue during which input and guidance from multiple stakeholders from around the world will be solicited before issuing a consensus study report.12 The Charo/Hynes committee represents the second step the NAS/NAM has taken on this issue following the highly successful and informative “International Summit on Human Gene Editing” held in December 2015 and chaired by Dr David Baltimore. This review touches on just a few of the issues that the committee will address in their more comprehensive study.
Germ line
Unintended germ line editing.
The ethical issues for unintended modification of germ cells in the course of an in vivo gene-editing therapy are not different from those for therapeutic approaches that would use gene addition. It has been a founding tenet of the field of human gene therapy that the human germ line in patients should not be perturbed genetically. Preclinical studies required for early clinical trials using in vivo viral-mediated gene delivery (eg, by adenovirus or AAV vectors) demonstrated that there was not permanent gene transfer to egg or sperm in rodents and large animal models.13-15 Similarly, it would be reasonable to demonstrate that a specific in vivo gene-editing approach would also not detectably modify germ cells in appropriate preclinical models as part of preclinical safety assessment to minimize risks of inadvertent modification of the germ line in patient’s germ cells. Of course, there are also risks of spontaneous germ line alterations in embryos due to the ex vivo manipulations per se, in addition to effects for the genome-editing reagents.
Intentional germ line editing.
The devastating nature of certain genetic diseases morally drives physicians and scientists to find ways of curing or preventing such diseases. One approach to these diseases, currently only theoretic, would be to use genome editing of germ cells or zygotes to correct disease-causing mutations in all cells of the person to curatively prevent these diseases from developing (Figure 2). Although the goal may be to correct the genetic defect in some or all of the somatic cells of the person, their germ cell–forming tissues (testes or ovaries) would also be gene modified and this modification could be passed in the germ line to successive generations. One of the inadvertent but accepted effects of the successes of modern medicine is that by providing improved survival it has indirectly altered the allele frequency of certain disease-causing mutations in the human gene pool. However, there is a higher ethical concern for the direct intentional alteration of the germ line to eliminate pathogenic mutations that may through childbearing introduce the modified genomes into the human gene pool.
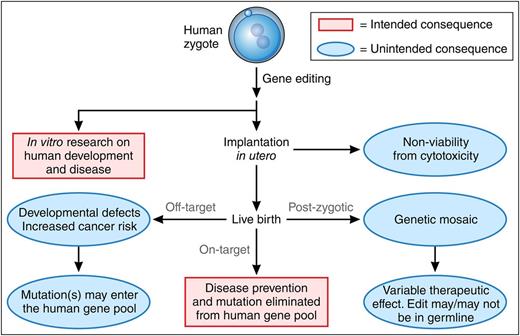
Consequences of gene editing in human zygotes. Intended outcomes appear in red boxes; unintended outcomes appear in blue type. Gene editing performed on the unicellular zygote may, in theory, be used clinically to prevent an inherited disorder by correcting a genetic defect. The resulting gene-corrected embryo could be implanted in utero for development to a live birth with every cell containing the genome edit. In theory, germ line correction of the mutation would lead to its elimination from the human gene pool. However, gene editing in human embryos to produce a live birth is consensually prohibited. In vitro research using gene editing in human zygotes may provide valuable information about human development and human disease mechanisms, which could lead to new treatments. However, several unintended consequences may occur from gene editing of human embryos, including direct cytotoxicity to the zygote from the manipulation; off-target editing and genotoxicity which could be pathogenic or be propagated to the human gene pool and may become homozygous in subsequent generations; or genetic mosaicism if gene modification occurs at a time after the zygote divides (postzygotic), leading to heterocellular gene-modification effects. Professional illustration by Patrick Lane, ScEYEnce Studios.
Consequences of gene editing in human zygotes. Intended outcomes appear in red boxes; unintended outcomes appear in blue type. Gene editing performed on the unicellular zygote may, in theory, be used clinically to prevent an inherited disorder by correcting a genetic defect. The resulting gene-corrected embryo could be implanted in utero for development to a live birth with every cell containing the genome edit. In theory, germ line correction of the mutation would lead to its elimination from the human gene pool. However, gene editing in human embryos to produce a live birth is consensually prohibited. In vitro research using gene editing in human zygotes may provide valuable information about human development and human disease mechanisms, which could lead to new treatments. However, several unintended consequences may occur from gene editing of human embryos, including direct cytotoxicity to the zygote from the manipulation; off-target editing and genotoxicity which could be pathogenic or be propagated to the human gene pool and may become homozygous in subsequent generations; or genetic mosaicism if gene modification occurs at a time after the zygote divides (postzygotic), leading to heterocellular gene-modification effects. Professional illustration by Patrick Lane, ScEYEnce Studios.
In addition to the issues of direct intentional editing of the germ line, other ethical considerations concerning germ line editing include the goal of the intervention: disease curative or “genetic enhancement” and whether the change will re-create what is naturally found in human genetics or whether it will create a human genome that is not normally found in the human population, such as by using editing to add a gene into a specific genomic location. Given that our understanding of the genome is still rudimentary, editing with intent to “improve” is fraught with many caveats, not the least of which is that we do not know, may never know, or nor even be able to define what an “improved” genome would look like.
Although there are moral arguments for and against germ line editing, there are 3 important practical limitations to germ line genome editing. The first is that there are only a tiny number of instances in which preimplantation genetic diagnosis could not achieve what one might want to achieve with germ line genome editing. Nonetheless, there are examples in which preimplantation genetic diagnosis would not work, most notably when couples who share a homozygous-recessive disease have children or when 1 parent is homozygous for an autosomal-dominant disease. The second is that current procedures of zygote editing, even in lower rodents, are still inefficient with multiple zygotes having to be manipulated and implanted in order to generate a mosaic genetically edited progeny. The third, and perhaps most problematic, practical limitation is that one cannot assure that a harmful unintended heritable modification does not occur in the context of an intentional germ line editing procedure, when there is no capacity for characterization of a modified germ cell postprocedure. Even with only the best of intentions to remove a disease allele, present technology cannot assure us that unintended modifications created through an editing procedure would not result in a devastating long-term outcome such as cancer or adverse developmental effects if one were to modify a zygote. These, perhaps insurmountable, safety concerns might be abrogated by editing germ line stem cells followed by deep sequencing to assure no unintended modifications have occurred. The transplantation of human germ line stem cells or the generation of human germ cells from germ line stem cells, even without modification, however, has not been performed.
Although modifying human zygotes and implanting them to generate humans perhaps has insurmountable safety and ethical limitations, the use of editing as a research tool to understand early human development or disease pathogenesis and treatment without implantation is probably ethically permissible based on current guidelines (for examples, see National Institutes of Health [NIH]16 ). The major ethical issue for the research use of editing in human zygotes is that it would require the specific generation of 1-cell zygotes for study as zygotes generated by in vitro fertilization are already at the multicellular blastocyst stage and thus beyond the very earliest stages of zygote development. Moreover, at the multicellular blastocyst stage, any editing approach would likely create a mosaic of edited cells in which different cells acquired different genomic changes, thus confounding any experimental results. In fact, the Human Fertilization and Embryology Authority in the United Kingdom has recently given permission to Kathy Niakan, a developmental biologist working at the Francis Crick Institute in London, to perform in vitro studies using clustered regularly-interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome manipulation in human embryos.17
Somatic cells
Modification of somatic cells by gene editing does not raise unique ethical issues above those raised by gene addition. Both approaches have ranges of efficacy and potential adverse effects, and their use is predicated on defining a risk-benefit ratio that favors benefit to the patient. For gene-editing procedures, as with any other medical procedure, this balance will be variably defined depending on the type of cell being edited (eg, terminally differentiated somatic cells vs stem cells), the nature and extent of the editing procedure (eg, number of loci being edited and their potential oncogenic activities), and the therapeutic application (eg, life-threatening disease vs enhancement/cosmetic). Gene-edited cells have been used in clinical trials for HIV-1, by zinc finger nuclease (ZFN)-mediated disruption of the CCR5 HIV coreceptor,18 and more recently for the treatment of leukemia through the use of allogeneic T cells armed with a chimeric antigen receptor, with transcription activator-like (TAL) effector nuclease (TALEN)-mediated editing to disrupt the endogenous T-cell receptor (TCR) gene to prevent alloreactivity.19
Some have raised the possibility that genome editing of somatic cells for “enhancement” poses a specific ethical issue. The ethical issue of enhancement by genome editing, however, is probably not ethically distinct from enhancement using other gene-therapy strategies, including the use of lentiviral vectors or AAV vectors and thus is a relatively mature ethical issue. We note that “enhancement” ranges from engineering a cell to overexpress a therapeutic protein, as was done using lentiviral modification of HSCs to treat metachromatic leukodystrophy,20 to using genome editing to knock out CCR5 to “enhance” resistance to HIV infection,18 to as-yet-unproven genetic modifications that might result in cosmetic changes (intelligence, muscular function, height, etc). As noted previously, given our still rudimentary knowledge of the complex and interacting genome, the issue of whether we know what an “improved” genome really is remains as the concept of unintended/unanticipated consequences comes to the fore. Nonetheless, any proposed gene-therapy approach aimed at enhancement must still pass the well-established threshold that the potential risk/benefit ratio is acceptable, informed consent is provided under local institutional review board guidance, and all required regulatory approvals are obtained from the relevant oversight entities (eg, the NIH Recombinant DNA Advisory Committee [RAC] and the Food and Drug Administration [FDA] in the United States, and the European Medicines Agency [EMA], etc, in the European Union).
Conclusions
Recent advances in editing technology have opened the door to application of editing technology to achieving a molecular level cure of many genetic diseases. Aside from moral considerations related to editing of the human germ line, at its present level of development, gene-editing technology cannot ethically be applied for therapeutically correcting disease genes in pluripotent/germ line cells due to our lack of knowledge of detrimental effects that might be attributable to editing procedures. In contrast, it is clear that gene-editing technology may be beneficially applied in somatic cell contexts where the risk-to-benefit ratio can be determined to be favorable to patients through rigorous safety and efficacy assessments.
Risk assessment and regulatory concerns for therapeutic gene editing
Assessment of genotoxicity
Risks associated with gene editing are summarized (Table 2). Of course, a major area of concern is potential genotoxic effects of gene editing. Although the understood risks of gene therapy using integrating vectors primarily relate to uncontrolled integrations causing an inadvertent activation of a proto-oncogene, producing an aberrant spliced gene product, or inactivating a tumor-suppressor gene, the primary genotoxic risk of nuclease-mediated genome editing is probably most related to the induction of DSBs in the genome.
Risks of genome editing
| Toxicity . | Potential risks . |
|---|---|
| Genotoxicity | (On-target) Local mutations at target locus: insertions/deletions, base substitutions; end capture of donor, 1 sided or nonhomologous recombination, and unintended gross chromosomal rearrangements; (off-target) mutations at sites other than target locus, including gross chromosomal rearrangements |
| Potential consequences: local gene inactivation, dysregulation, trans-activation; deletions, inversions, translocations; transformation, cell cycle disruptions, apoptosis | |
| Nuclease related | Off-target endonuclease activity |
| Persistent endonuclease activity (off-target mutations and/or activation of the DNA damage response) | |
| Immunogenicity | |
| Vector related (nucleic acids, nucleoproteins, nanoparticles, viral vectors) | Random integration of donor vector (illegitimate recombination at a spontaneous or induced DSB) leading to local gene inactivation, dysregulation, trans-activation |
| Random integration of nuclease expression vector (illegitimate recombination at a spontaneous or induced DSB) leading to persistent nuclease expression | |
| Delivery related (transfection, microinjection, electroporation, viral vector) | Acute cytotoxicity |
| Induction of inflammatory responses to vector components | |
| Delivery device related (needles, catheters, transplants, stereotactic, electroporation in vivo) | Tissue injury/inflammation |
| Infection | |
| Mechanical mishaps | |
| Ex vivo cell processing related | Cell survival |
| Microbial contamination | |
| Loss of stem cell activity and/or engraftment activity; impaired differentiation/function; inherent genome instability during culture |
| Toxicity . | Potential risks . |
|---|---|
| Genotoxicity | (On-target) Local mutations at target locus: insertions/deletions, base substitutions; end capture of donor, 1 sided or nonhomologous recombination, and unintended gross chromosomal rearrangements; (off-target) mutations at sites other than target locus, including gross chromosomal rearrangements |
| Potential consequences: local gene inactivation, dysregulation, trans-activation; deletions, inversions, translocations; transformation, cell cycle disruptions, apoptosis | |
| Nuclease related | Off-target endonuclease activity |
| Persistent endonuclease activity (off-target mutations and/or activation of the DNA damage response) | |
| Immunogenicity | |
| Vector related (nucleic acids, nucleoproteins, nanoparticles, viral vectors) | Random integration of donor vector (illegitimate recombination at a spontaneous or induced DSB) leading to local gene inactivation, dysregulation, trans-activation |
| Random integration of nuclease expression vector (illegitimate recombination at a spontaneous or induced DSB) leading to persistent nuclease expression | |
| Delivery related (transfection, microinjection, electroporation, viral vector) | Acute cytotoxicity |
| Induction of inflammatory responses to vector components | |
| Delivery device related (needles, catheters, transplants, stereotactic, electroporation in vivo) | Tissue injury/inflammation |
| Infection | |
| Mechanical mishaps | |
| Ex vivo cell processing related | Cell survival |
| Microbial contamination | |
| Loss of stem cell activity and/or engraftment activity; impaired differentiation/function; inherent genome instability during culture |
There are risks for on-target DSBs at the target locus to either disrupt the gene or cause chromosomal rearrangements if it interacts with spontaneous nonnuclease DSBs elsewhere in the genome. Thus, assessment of the potential genotoxic risk of the on-target DSB should be part of a safety package. Much more debated are the issues of off-target DSBs (DSBs generated from the engineered nuclease at unintended sites). These may result in the creation of insertions/deletions in a genetic element, such as protein coding gene, thereby inactivating it, or the creation of gross chromosomal rearrangements through the illegitimate recombination between 2 DSBs, either both created by the engineered nuclease or 1 created by the engineered nuclease and 1 occurring spontaneously. Thus, assays that can assess both of these types of events should be used in assessing the preclinical safety of a given editing process (Table 3).21-33 In addition to assessing the genotoxicity of the nucleases, if methods are used to perturb how the DSB is repaired in an attempt to increase HR-mediated editing (eg, inhibition of NHEJ), these manipulations could also lead to increased genome instability and would need to be carefully assessed.
Preclinical safety assays to detect and define the genotoxic effects of nucleases and homology donors
| Class of assay . | Methods . | Advantages . | Disadvantages . |
|---|---|---|---|
| Sequence based | Genome-wide sequencing | Unbiased assessment of entire genome | Cannot be used on populations of cells because of sensitivity |
| Background error rate remains high | |||
| Bioinformatics based: use computational tools to predict off-target sites and then deep sequence predicted sites to assess whether insertions/deletions are being created | Many currently available algorithms and sequencing platforms make this easily available | Prediction algorithms have not routinely identified all of the off-target sites that have been found using other methods | |
| Karyotyping | Routine procedure | Only assesses a small number of cells (20-50) | |
| Unbiased DSB capture methods combined with sequencing; can be done by capturing break with plasmid DNA,21 AAV,22 IDLV,23,24 or oligos,25 or by directly sequencing sites of DSBs (Digenome Seq26 or BLESS27 ). | Unbiased | Technically difficult with sensitivity of 0.1% | |
| Translocation capture28 | Unbiased assessment for genome rearrangements | Technically difficult; primarily assesses rearrangements from the intended target site not between 2 off-target sites | |
| “Onco-chip” | Specifically assesses for mutations in genes associated with cancer | Low sensitivity; list may be incomplete | |
| Function based | Staining for DSBs (53BP129 or gH2AX30 ) | Simple and quantitative and can be applied to a wide variety of cell types and used in vivo | No information on sites of DSBs or consequence of DSBs |
| Relative cell proliferation29 | Simple and quantitative and can be applied to a wide variety of cell types | Does not give information on mechanisms | |
| Cell cycle perturbations30 | Quantitative measure of functional effect on key cell function | Only used in a specialized cancer cell line | |
| Clonal dynamics assays31 | Can assess whether nucleases can induce changes in clonal representation in large populations of cells | Requires cell population to be grown for long periods of time; could be confounded by the integration site of barcode to mark clones | |
| Colony-forming replating assay32 | Useful assessment for retroviral and lentiviral vectors | Not clinically validated and has not yet been adopted to genome-editing strategies | |
| Lineage reconstitution and colony-forming assays5 | Direct functional measure of edited cell’s behavior | Sensitivity to important genotoxic events is not validated | |
| In vitro transformation33 | The best current model for assessing cell transformation | Very insensitive and not validated for predicting human |
| Class of assay . | Methods . | Advantages . | Disadvantages . |
|---|---|---|---|
| Sequence based | Genome-wide sequencing | Unbiased assessment of entire genome | Cannot be used on populations of cells because of sensitivity |
| Background error rate remains high | |||
| Bioinformatics based: use computational tools to predict off-target sites and then deep sequence predicted sites to assess whether insertions/deletions are being created | Many currently available algorithms and sequencing platforms make this easily available | Prediction algorithms have not routinely identified all of the off-target sites that have been found using other methods | |
| Karyotyping | Routine procedure | Only assesses a small number of cells (20-50) | |
| Unbiased DSB capture methods combined with sequencing; can be done by capturing break with plasmid DNA,21 AAV,22 IDLV,23,24 or oligos,25 or by directly sequencing sites of DSBs (Digenome Seq26 or BLESS27 ). | Unbiased | Technically difficult with sensitivity of 0.1% | |
| Translocation capture28 | Unbiased assessment for genome rearrangements | Technically difficult; primarily assesses rearrangements from the intended target site not between 2 off-target sites | |
| “Onco-chip” | Specifically assesses for mutations in genes associated with cancer | Low sensitivity; list may be incomplete | |
| Function based | Staining for DSBs (53BP129 or gH2AX30 ) | Simple and quantitative and can be applied to a wide variety of cell types and used in vivo | No information on sites of DSBs or consequence of DSBs |
| Relative cell proliferation29 | Simple and quantitative and can be applied to a wide variety of cell types | Does not give information on mechanisms | |
| Cell cycle perturbations30 | Quantitative measure of functional effect on key cell function | Only used in a specialized cancer cell line | |
| Clonal dynamics assays31 | Can assess whether nucleases can induce changes in clonal representation in large populations of cells | Requires cell population to be grown for long periods of time; could be confounded by the integration site of barcode to mark clones | |
| Colony-forming replating assay32 | Useful assessment for retroviral and lentiviral vectors | Not clinically validated and has not yet been adopted to genome-editing strategies | |
| Lineage reconstitution and colony-forming assays5 | Direct functional measure of edited cell’s behavior | Sensitivity to important genotoxic events is not validated | |
| In vitro transformation33 | The best current model for assessing cell transformation | Very insensitive and not validated for predicting human |
BLESS, breaks labeling, enrichment on streptavidin, and next-generation sequencing; IDLV, integration-deficient lentiviral vector; oligo, oligonucleotide.
An important consideration is that as editing becomes easier, there is the possibility of editing 2 or more loci simultaneously (“multiplexing”).34 If multiplexing is to be applied in the clinical setting, toxicity assays should be designed to assess whether it creates heightened risks by simultaneously creating multiple DSBs that may interact, yielding translocations. Although the assessment of genotoxic risk of nuclease-mediated genome editing should be designed around the potentially pathologic DSB, there is currently no reason to alter the set of assays used based on the nuclease platform used, whether it be ZFNs, TALENs, CRISPR/Cas9, variants thereof, or some other nuclease platform yet to be discovered.
There are at least 4 important caveats to this genotoxic risk assessment. The first is that the genome is undergoing ongoing spontaneous mutagenesis with gross chromosomal rearrangements continuously. Thus, the genotoxic risk of the editing process (both from the nuclease itself and from the cell manufacturing process) should be put into the context of the natural, ongoing genomic mutations that are occurring in cells all of the time. It is estimated, for example, that in healthy stem cells in vivo, 1 to 3 mutations are occurring per cell division. The measurement of the mutation rate in stem cells ex vivo in a superphysiologic oxygen environment under stressful culture conditions is not known and should be an active area of study. The second caveat is that current sequencing-based genotoxicity assays, whether directed to specific sites or unbiased, have a technical limit of detection of ∼1 in 10 000. Thus, when creating a modified cell population of several hundred million cells, even with the most sensitive sequencing-based assays, there could still be tens of millions of cells that have undetected nuclease-induced off-target mutations and rearrangements. The third is that although whole-genome sequencing might seem to be an ideal method to assess genotoxicity, this approach will only work in a therapeutic process in which transplanted cells are derived from a single clone. Moreover, even whole-genome sequencing has a low sensitivity and thus might miss oncogenic mutations that spontaneously occur as a therapeutically relevant number of cells are generated from a single cell during the expansion process, and not from the gene editing per se. The fourth is the recognition that a large number of routine therapies cause genotoxicity, even in cells that are not the target of the therapy. For example, the only current cure for sickle cell disease is allogeneic HSC transplantation and it involves exposing all the cells of the patient to genotoxic agents. Thus, “genotoxicity” itself is not an absolute contraindication, but instead, as discussed earlier, must be considered as part of a standard risk/benefit analysis.
Therefore, sequencing-based methods of analysis should be complemented by functional methods of assessing genotoxicity (Table 3). Functional genotoxicity assays would measure various aspects of cellular behavior including apoptosis, proliferation, differentiated functionality (such as the ability to perform equivalently to unmodified cells), and potential to transform. Although important information can be gained using these assays in surrogate cell types, such as transformed cell lines, which can then guide how to improve the relative specificity, the key assays need to be performed in the therapeutic cell type of interest. Apoptosis, proliferation, and functionality assays can often be performed in the relevant cell type of interest, but there are no established functional validated assays for transformation of primary human stem cells (other than teratomas from pluripotent stem cells). Moreover, there are many types of human cancers, including human leukemias, which simply do not grow in immunodeficient mouse models. Thus, assays such as fibroblast soft-colony-forming assays and leukemia formation assays in immunodeficient mice, may simply be irrelevant to truly understanding the functional genotoxic risks of any gene-therapy process, including nuclease-mediated genome editing. Given these caveats on the lack of validated assays, it might be prudent in some of the first genome-editing clinical trials to demonstrate at a minimum that the production of DSBs is not detectable in genes that are associated with human cancers by whichever assay is used. Such panels of genes have already been generated to study the genetics of cancer and could be repurposed to assess the potential genotoxicity of genome editing. Again, such panels have only a limited sensitivity and are currently not designed to detect mutation frequencies in a population much below 1%.
For genome editing, one class of safety assays directly evaluates the genotoxicity of the editing process. The assays of genotoxicity should be relevant to the editing process itself and not be generic in nature unless required by formal laws and/or regulations. That is, assays that assess genotoxic risk of integrating viruses or small molecules may or may not be relevant to assessing the genotoxic risks of engineered nuclease-mediated genome editing. One unfortunate key point in the assessment of genotoxicity, however, is that there are no established preclinical assays that have been validated to predict genotoxic risk in human clinical trials. This unfortunate reality not only applies to genome editing but also to other forms of gene therapy including the use of AAV and lentiviral vectors. The ability to detect rare events is limited in the relatively low numbers of cells and relatively short time periods of examination that are assessed in murine models compared with clinical-scale numbers of cells and lifetime at risk.
The regulatory framework for gene-editing procedures will necessarily focus on evaluating the potential medical benefit of the procedure vs the potential for toxicity. The genotoxic risk assessment probably should also be tailored to whether the approach is to be ex vivo vs in vivo. Although there are certain shared risks for both approaches, including potential immune reactions to the nuclease component (ZFNs, TALENs, and CRISPR/Cas9 all use proteins or peptides components that are derived from bacteria), the potential immunogenicity for an in vivo approach is likely higher. Moreover, it is currently easier to limit the duration of the nuclease expression in an ex vivo approach, such as by delivering the nuclease as protein or messenger RNA (mRNA), which not only limits the potential immune response but also limits the duration of exposure of the genome to a DSB-creating agent. Thus, safe in vivo strategies should be designed such that the duration of nuclease expression is limited to minimize the exposure and potential genome modification of both target cells and nontarget cells. AAV vectors, for example, have been shown to transduce multiple different cell types and to deliver sustained expression of the transgene it carries for years.35 Thus, if a nuclease is delivered on an AAV vector in vivo that can both transduce target cells and nontarget cells, a method needs to be developed to turn off the nuclease so it does not create ongoing, perhaps low-grade, genotoxic DNA damage in target and nontarget cell types.
There may be a “regulatory learning curve” such that as experience is gained with particular nuclease platforms, similar ones targeting other loci may require less extensive preclinical assessment. One way to potentially streamline regulatory reviews would be to establish a safety information “master file” that can then be reused in subsequent proposed therapeutic applications. Given that there are both relatively specific and nonspecific nucleases from all of the major nuclease platforms, it is likely that such master files would be related to a specific nuclease rather than applied more broadly to a class of nuclease. As assays and technologies improve, it would be important to update the nuclease master file appropriately. Similarly, once a defined genomic target is established to be a safe harbor for targeted transgene addition by human genetics, functional genomics, and functional phenotype assays, that site should again have a “master file” that can be used for subsequent therapeutic purposes. Thus, it would be ideal to have master files on both specific nucleases and specific sites that could be accessible and broadly used as investigators develop different therapeutic genome-editing applications. In such subsequent applications, therefore, safety assessment could be focused on the type of edit generated, including the potential safety of a transgene being expressed from that locus.
One of the potential advantages of genome editing as compared with using integrating viral vectors is the reduction of uncontrolled integration events. But, it should be noted that whenever a DNA molecule is introduced into a cell, whether as a naked piece of DNA, as an AAV vector, or even after integration-defective lentiviral transduction, that DNA can integrate into the genome through illegitimate recombination, sometimes directly into the nuclease-induced DSB and other times into spontaneous DSBs. Thus, assessment of the potential genotoxicity from illegitimate recombination causing uncontrolled integration should also be part of the safety assessment, particularly when the nuclease is expressed from a DNA vector. Delivering the nuclease as mRNA or protein, either for in vivo or ex vivo approaches, would obviate that potential risk for the nuclease.
Assessment of safety of the cell
Like all gene-therapy approaches, a safety evaluation of the genetically modified cell phenotype is critical. Thus, whether one uses genome editing or lentiviral delivery or some other form of gene therapy to generate a modified cell, the safety phenotype of the cell or cell population must be assessed, irrespective of how it was generated. For example, if one generates a genetically modified T cell with a new checkpoint defect by any mechanism, the safety of checkpoint-deleted T cells needs to be assessed; for example, will they cause uncontrolled autoimmunity, clonal expansion, etc?
A unique situation arises for application of gene-editing procedures to correct genetic mutations with well-defined causal roles in a particular genetic disease. In these cases, the “potency” or “therapeutic benefit” of a corrected allele may be inferred because of the effectiveness of the wild-type allele, and the overall benefit of the gene-editing procedure will devolve to an assessment of the efficiency of engraftment of the repaired cell population and the safety of the editing process.
Importantly, gene correction is an important area for development of a rationale regulatory framework, as these applications will enable personalized or near-personalized curative therapies for patients suffering from diseases caused by rare genetic mutations. In such a context, extensive evaluation for safety and efficacy may not be possible, and regulatory practice will require modification to account for the use of an approved “editing procedure” in which different editing reagents can be qualified to a standard, and then substituted without requirement for a subsequent full regulatory review. In this way, it might be possible to establish a regulatory path for genome editing to treat N = 1 diseases and thus open the great potential of therapeutic genome editing to a much wider range of diseases. One of the features of genome editing is that it targets the same relatively homogeneous and polymeric DNA molecule (in contrast to small-molecule drugs that target heterogeneous proteins). Thus, the chemical nature of the target does not change substantially. Moreover, within a given platform, even 2 nucleases that target different genomic sites are chemically nearly identical. These features also might support the development of a streamlined regulatory path.
It is not possible to simply list a series of safety assessments that need to be performed and the cutoff values that would allow a protocol to proceed to the clinic. Rather, safety assessments need to be done that are relevant, quantitative, and controlled, to define the baseline properties of the approach, which can then be compared with clinical outcomes.
Summary
Genome editing holds great promise to provide a precise set of tools for counteracting genetic diseases. But, as Spiderman cautions, “With great power, there must come great responsibility.” Moving these methods to clinical applications must proceed judiciously with efforts made to define risks a priori and minimize them under appropriate regulatory oversight.
Authorship
Contribution: D.B.K., M.H.P., and A.M.S. contributed equally to the writing of this paper.
Conflict-of-interest disclosure: D.B.K. holds equity in Orchard Therapeutics and receives compensation for consulting for them. M.H.P. holds equity in CRISPR Therapeutics and receives compensation for consulting to them. A.M.S. holds equity in bluebird bio and receives compensation for consulting for them.
Correspondence: Donald B. Kohn, David Geffen School of Medicine, University of California, Los Angeles, 3163 Terasaki Life Science Building, 610 Charles E. Young Dr E, Los Angeles, CA 90097; e-mail: dkohn1@mednet.ucla.edu.
References
Author notes
D.B.K., M.H.P., and A.M.S. contributed equally to this study.