Abstract
HIV/AIDS has long been at the forefront of the development of gene- and cell-based therapies. Although conventional gene therapy approaches typically involve the addition of anti-HIV genes to cells using semirandomly integrating viral vectors, newer genome editing technologies based on engineered nucleases are now allowing more precise genetic manipulations. The possible outcomes of genome editing include gene disruption, which has been most notably applied to the CCR5 coreceptor gene, or the introduction of small mutations or larger whole gene cassette insertions at a targeted locus. Disruption of CCR5 using zinc finger nucleases was the first-in-human application of genome editing and remains the most clinically advanced platform, with 7 completed or ongoing clinical trials in T cells and hematopoietic stem/progenitor cells (HSPCs). Here we review the laboratory and clinical findings of CCR5 editing in T cells and HSPCs for HIV therapy and summarize other promising genome editing approaches for future clinical development. In particular, recent advances in the delivery of genome editing reagents and the demonstration of highly efficient homology-directed editing in both T cells and HSPCs are expected to spur the development of even more sophisticated applications of this technology for HIV therapy.
Introduction
HIV-1 infection presents a unique set of challenges for the development of effective therapies. During infection, the virus inserts a copy of itself into the genome of a cell and thereby becomes a permanent part of the cell’s genetic material. On occasion, expression from such integrated viruses is suppressed, leading to a latent viral state that is not impacted by the antiretroviral drug therapies (ARTs) that target replicating viruses. Although rare,1 such latently infected cells comprise a significant reservoir in the body with a long half-life. Moreover, latent virus can be reactivated in response to changes in a cell’s activation status, which leads to a rapid rebound of viremia if ART is discontinued.2-4 As a result, HIV management requires life-long ART, which does not ever cure people.
Adherence to a daily ART regimen can be challenging. Compliance is impacted by accessibility, drug-related side effects, lack of health literacy, and social and emotional challenges. Consequently, a significant portion of patients on ART do not achieve and maintain full virologic control, and even in a developed country with highly accessible ART such as the United States, only a third of the estimated 1.2 million people currently infected with HIV-1 achieve optimal ART.5 HIV infection, even when fully suppressed, can continue to impact immune function, in particular as a result of the decimation of CD4+ T cells that occurs during the acute initial phase of infection. Indeed, some patients never recover normal levels of CD4+ T cells, even when ART provides effective virologic control.6
The limitations of ART and the similarities between HIV infection and a more traditional genetic disease made HIV/AIDS an early poster child for the development of experimental genetic-, cellular-, and immune-based therapies.7-9 Despite the recognized complexity of such approaches, the possibilities of one-shot treatments that could both restrict viral replication and restore immune function are spurring development. To date, most gene therapy approaches for HIV have involved the addition of anti-HIV genes to protect CD4+ T cells from infection. By reducing the number of susceptible cells, such strategies are expected to reduce overall viremia, while the selective survival of the engineered cells would increase the effectiveness of the treatment over time. In addition, the direct protection of CD4+ T cells could restore immune function, allowing the body to fight back against both the symptoms of the infection and the virus itself. A virtuous cycle is thus envisioned.
Several anti-HIV genes have already been tested in this way in clinical trials, including dominant negative viral proteins (Rev M10) and peptides (C46), viral RNA decoys (TAR and RRE), and RNA-based methods to suppress either host or viral genes using ribozymes, antisense RNA, and RNA interference technologies.10-16 The candidate anti-HIV factors are delivered ex vivo to either CD4+ T cells or their in vivo precursors: the hematopoietic stem and progenitor cells (HSPCs). To achieve permanent gene expression, integrating retroviral and lentiviral vectors are typically used, taking advantage of viral machineries that have evolved to permanently integrate into the genomes of hematopoietic cells. On the down side, these vectors can suffer from lack of long-term gene expression and genotoxic risk due to random genomic integration.17,18 Clinical trials to date have established a portfolio of candidate anti-HIV genes, demonstrated safety, and created an appetite for gene therapy, but stopped short of demonstrating efficacy.
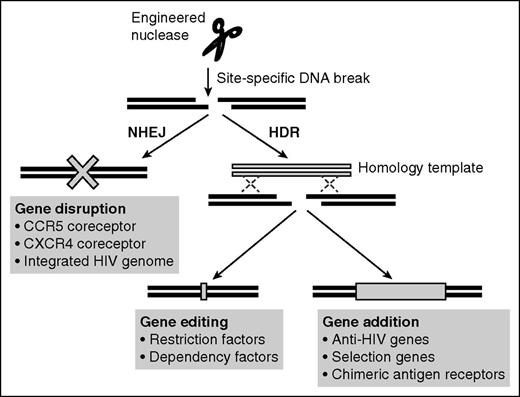
Genome editing has emerged recently as a more precise way to engineer cells that could also be applied to HIV/AIDS. At its heart, the technology uses engineered nucleases to introduce DNA double-strand breaks (DSBs) at a targeted locus, whose subsequent repair is exploited to achieve different outcomes. If the nonhomologous end joining (NHEJ) repair pathway is accessed, the end result can be random insertions/deletions (indels) at the break site that result in gene disruption. Alternatively, more precise repair pathways based on homologous recombination can be hijacked to copy information from an introduced DNA homology template. Such homology-directed repair (HDR) can promote a specific gene editing/mutation event or allow the site-specific addition of larger gene cassettes at the break site. The initial DSB generation can be achieved using different engineered nuclease platforms, including homing endonucleases, zinc finger nucleases (ZFNs), transcription activator–like effector nucleases (TALENs), and Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9.19,20 As described below, all 3 of the possible outcomes of DSB repair are being developed for HIV-1 treatment (Figure 1).
Potential applications of genome editing in anti-HIV therapy. Site-specific DNA breaks created by engineered nucleases can be repaired by error-prone NHEJ or, if a homologous DNA repair template is also present, by more precise HDR pathways. NHEJ frequently results in indels, allowing disruption of genes such as the HIV-1 coreceptors CCR5 and CXCR4 or integrated HIV-1 genomes. HDR could be used to introduce small mutations into host restriction factors to restore anti-HIV activities or into host dependency factors to limit their use by the virus. Alternatively, HDR could be used to site-specifically insert anti-HIV genes at the site of the DNA break, including at a disrupted CCR5 locus.
Potential applications of genome editing in anti-HIV therapy. Site-specific DNA breaks created by engineered nucleases can be repaired by error-prone NHEJ or, if a homologous DNA repair template is also present, by more precise HDR pathways. NHEJ frequently results in indels, allowing disruption of genes such as the HIV-1 coreceptors CCR5 and CXCR4 or integrated HIV-1 genomes. HDR could be used to introduce small mutations into host restriction factors to restore anti-HIV activities or into host dependency factors to limit their use by the virus. Alternatively, HDR could be used to site-specifically insert anti-HIV genes at the site of the DNA break, including at a disrupted CCR5 locus.
CCR5 disruption: the first-in-human application of targeted nucleases
It is no accident that the first application of targeted nucleases in humans was an anti-HIV strategy based on preventing expression of the CCR5 gene. Achieving gene disruption is far simpler than gene editing or insertion, because it relies only on the activity of the nuclease and does not require the additional delivery of a homology template. In addition, NHEJ is active throughout the cell cycle, whereas HDR is mainly restricted to S and G2, when repair templates are available in the form of sister chromatids.21,22 Finally, CCR5 represents a uniquely attractive disruption target, being a nonessential human gene whose absence provides potent anti-HIV protection.
CCR5 is the entry coreceptor used by the majority of HIV strains and in particular by the transmitting and early infecting strains.23 The CCR5Δ32 allele contains a 32-bp deletion that results in a truncated protein that is not expressed on the cell surface. The allele confers protection against HIV-1 infection without adverse health consequences in homozygotes, which constitute ∼1% of the Caucasian population, and HIV-infected heterozygotes exhibit delayed disease progression.24-26
The potential benefit of a CCR5 targeted gene therapy was strikingly demonstrated in 2009, in the first and only reported case of an HIV cure. The “Berlin patient” received an allogeneic HSPC transplant from a CCR5Δ32 donor during treatment of acute myeloid leukemia and has remained HIV-1 free without ART since then.27,28 Although he received significant conditioning as part of his treatment and developed graft-versus-host-disease, both of which likely reduced his HIV-1 reservoir, the CCR5Δ32 genotype of his donor is considered an essential component of his cure, because other attempts using HSPC transplants from CCR5 wild-type donors have not been curative.29,30
The extraordinary finding of a single individual cured of HIV-1 galvanized efforts to use gene therapy to create CCR5-negative autologous T cells or HSPCs in HIV-infected patients, with the goal of providing the same curative benefits without the challenges of allogeneic transplantation. At the same time, genome engineering emerged as a preferred method to achieve complete and permanent CCR5 depletion,31 which did not suffer from the incomplete or nonpermanent effects of protein- and RNA-based strategies.32-36 However, the clinical implementation of genome engineering required considerable development efforts to identify protocols that could deliver engineered nucleases to target cells and maximize bi-allelic disruption, while limiting off-target effects at nontargeted sites. This entirely new class of gene therapy technologies also required the development of appropriate in vitro and in vivo model systems to allow the necessary and rigorous preclinical safety studies.37 Much like it has in the past for gene therapy, HIV disease has been a pioneer in these endeavors.
ZFN-mediated CCR5 disruption in CD4+ T cells
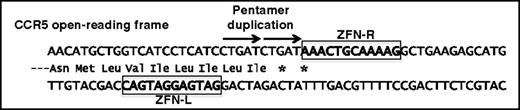
The first clinical trials to evaluate genome engineered CCR5-negative cells used ZFNs and built on prior experiences in T-cell adoptive transfer. A ZFN pair was identified that introduced a DSB at approximately nucleotide 160 of the CCR5 open-reading frame, corresponding to a site in the first of 7 transmembrane domains in the protein.38 The most frequent outcome of treatment with this ZFN pair is a 5 nucleotide addition, which accounts for ∼25% of all modified alleles.38,39 This pentamer duplication introduces 2 in-frame stop codons that block expression and also serves as a convenient genetic marker for estimating the overall frequency of CCR5 modification (Figure 2).
ZFN-mediated disruption of the CCR5 open-reading frame. A ZFN pair that binds CCR5 sequences (left and right, boxed) results in a DNA break whose repair most frequently results in a 5-bp duplication (arrowed) that introduces premature stop codons (*) into the open-reading frame.
ZFN-mediated disruption of the CCR5 open-reading frame. A ZFN pair that binds CCR5 sequences (left and right, boxed) results in a DNA break whose repair most frequently results in a 5-bp duplication (arrowed) that introduces premature stop codons (*) into the open-reading frame.
The anti-HIV efficacy of the ZFN pair was first demonstrated in preclinical studies using primary CD4+ T cells.38 A chimeric Ad5/F35 adenoviral vector was used to deliver the ZFN pair and was found capable of disrupting up to 40% to 60% of total CCR5 alleles. Furthermore, 33% of modified cells were disrupted at both alleles and therefore were effectively CCR5 null. In a mouse xenotransplantation model, HIV-1 challenge was seen to increase the proportion of modified CCR5 alleles threefold, confirming the expected survival advantage for the gene-edited cells. Mice receiving CCR5 ZFN-treated cells also better preserved their human CD4+ T cells and had lower viremia following HIV-1 challenge.38 Subsequently, Ad5/F35 delivery of this ZFN pair to CD3/CD28-stimulated CD4+ T cells was adapted for clinical scale, allowing the production of >1010CCR5-edited cells and paving the way for phase 1 clinical studies.40
The first-in-human genome editing trial to treat HIV (#NCT00842634) was initiated in 2009, primarily to evaluate the safety of modifying autologous CD4+ T cells in HIV-1–infected individuals41,42 (Table 1). Twelve patients on ART, with undetectable viral loads, were enrolled in 2 cohorts, based on whether their CD4+ T-cell counts were >450/mm3 (cohort 1) or at less optimal levels of 200 to 500/mm3 (cohort 2). Each participant received a single infusion of 5 to 10 billion ZFN-modified autologous CD4+ T cells. The infusions were well tolerated, with only 1 individual experiencing a minor infusion-related adverse event.
Clinical trials of HIV-infected individuals using ZFN-modified autologous T cells and HSPCs
| Clinical trial . | Status . | Cohorts/study populations . | Cell type . | ZFN delivery method . | Conditioning regimen . |
|---|---|---|---|---|---|
| NCT00842634 phase 1 | 2009-2013 Completed41 | On ART, aviremic, CD4 counts >450 | CD4 T cells | Adenovirus | None |
| On ART, aviremic, CD4 counts 200-500, ATI | |||||
| NCT01044654 phase 1 | 2009-2014 Completed | On ART, aviremic, CD4 counts 200-500 | CD4 T cells | Adenovirus | None |
| On ART, aviremic, CD4 counts >500, and CCR5Δ32 heterozygotes, ATI | |||||
| NCT01252641 phase 1/2 | 2010-2015 Completed | Not on ART, CD4 counts >500 | CD4 T cells | Adenovirus | None |
| NCT01543152 phase 1 | 2011-2016 Recruiting | On ART, aviremic, CD4 counts >500, ATI | CD4 T cells, and CD4/CD8 T cells | Adenovirus | Cytoxan (dose escalation) |
| NCT02225665 phase 1/2 | 2014-2018 Active, not recruiting | On ART, aviremic, CD4 counts >500, ATI | CD4/CD8 T cells | mRNA | Cytoxan (1 g/m2) |
| NCT02388594 phase 1 | 2015-2017 Recruiting | On ART, aviremic, CD4 counts >450, ATI | CD4 T cells | mRNA | Cytoxan (1 g/m2) |
| NCT02500849 phase 1 | 2015-2018 Recruiting | On ART, aviremic, CD4 counts 200-500, ATI | CD34 HSPC | mRNA | Busulfan (dose escalation) |
| Clinical trial . | Status . | Cohorts/study populations . | Cell type . | ZFN delivery method . | Conditioning regimen . |
|---|---|---|---|---|---|
| NCT00842634 phase 1 | 2009-2013 Completed41 | On ART, aviremic, CD4 counts >450 | CD4 T cells | Adenovirus | None |
| On ART, aviremic, CD4 counts 200-500, ATI | |||||
| NCT01044654 phase 1 | 2009-2014 Completed | On ART, aviremic, CD4 counts 200-500 | CD4 T cells | Adenovirus | None |
| On ART, aviremic, CD4 counts >500, and CCR5Δ32 heterozygotes, ATI | |||||
| NCT01252641 phase 1/2 | 2010-2015 Completed | Not on ART, CD4 counts >500 | CD4 T cells | Adenovirus | None |
| NCT01543152 phase 1 | 2011-2016 Recruiting | On ART, aviremic, CD4 counts >500, ATI | CD4 T cells, and CD4/CD8 T cells | Adenovirus | Cytoxan (dose escalation) |
| NCT02225665 phase 1/2 | 2014-2018 Active, not recruiting | On ART, aviremic, CD4 counts >500, ATI | CD4/CD8 T cells | mRNA | Cytoxan (1 g/m2) |
| NCT02388594 phase 1 | 2015-2017 Recruiting | On ART, aviremic, CD4 counts >450, ATI | CD4 T cells | mRNA | Cytoxan (1 g/m2) |
| NCT02500849 phase 1 | 2015-2018 Recruiting | On ART, aviremic, CD4 counts 200-500, ATI | CD34 HSPC | mRNA | Busulfan (dose escalation) |
Information obtained from website Clinicaltrials.gov and publically reported by sponsors.
In terms of safety, the CCR5 ZFN-modified cells displayed normal characteristics, including engraftment in all patients, persistence for ≥42 months after infusion, and normal trafficking to the rectal mucosa. The trial also attempted to discern any anti-HIV effects. Four weeks after infusion, cohort 1 patients underwent a 12-week analytical treatment interruption (ATI), during which ART was stopped. This usually results in rapid viral rebound within 2 to 4 weeks and the establishment of a plasma viral load set point that is similar to historical levels in each patient before ART initiation. Changes in either the time to rebound or the viral load set point could indicate an anti-HIV effect. In addition, allowing a limited period of HIV replication during ART cessation was hypothesized to enrich for the CCR5-negative, HIV-resistant cells.
In the 4 patients that completed ATI, HIV rebounded and reached a peak viremia during ART withdrawal. Although CD4+ T cell counts decreased in all patients during this period of viremia, the mean rate of decline of the CCR5-modified CD4+ T cells was slower than that of the unmodified cells (−1.81 vs −7.25 cells/mm3 per day), suggesting a protective effect of the CCR5 modifications. Notably, 1 patient had a particularly unusual response, showing both delayed viral rebound (6 weeks into ATI) and a peak viremia that was lower than the patient’s historical set point. Intriguingly, this was followed by a decrease in plasma HIV levels to below the limit of detection, before ART was resumed as planned. Subsequently, this patient was identified as being heterozygous for CCR5Δ32, leading to the hypothesis that this “halfway there” genotype had amplified the effects of the ZFN treatment.
Since this initial demonstration of clinical safety, subsequent trials have been designed to further optimize the treatment (Table 1). The parameters being evaluated include varying the input dose of cells, using multiple infusions of cells, including ZFN-modified CD8+ T cells in the graft, assessing the ability of Cytoxan to transiently reduce T-cell numbers and thereby improve engraftment of the infused T cells, and switching from adenoviral vector delivery of the ZFNs to using mRNA electroporation.
The possible impact of the heterozygous CCR5Δ32 genotype is also being assessed in a cohort of 10 patients in trial #NCT01044654. These patients underwent a 16-week ATI 2 months after infusion, during which time 3 patients were able to reduce and maintain viral loads at low (<1000 copies/mL) or undetectable levels. It has also been reported that 1 of the 3 patients maintained a viral load of <500 copies/mL, with CD4+ T-cell counts of >1000 cells/mL, for >1 year without ART.43,44 Long-term follow-up will determine whether a “functional cure” has been achieved in this patient.
A major concern for any clinical application of engineered nucleases is the potential for off-target genome modifications, reviewed elsewhere in this series. For the CCR5 ZFN pair in clinical development, a disruption rate of 5.39% has been observed in CD4+ T cells at the top predicted off-target site, the highly related CCR2 gene, under conditions that gave 36% on-target CCR5 disruption.38 The prediction, assessment, and mitigation of off-target activities have undergone major improvements in the last few years, so that if such off-target disruption at CCR2 proved to be problematic, it could be engineered around by selecting and developing a different ZFN pair.45,46 Of note, CCR2 disruption is predicted to be well tolerated and may even confer an additional anti–HIV-1 benefit, because a naturally occurring CCR2 mutant allele is associated with delayed progression to AIDS.47
Prospects for CCR5 disruption in stem cells
Engineering HSPCs instead of CD4+ T cells has the potential to provide a long-lasting source of modified cells and to additionally protect the CD4+ myeloid cells that are also susceptible to HIV-1. We previously demonstrated that the same CCR5 ZFN pair used in the T-cell trials could effectively modify CD34+ HSPCs, isolated from cord blood, fetal liver, or mobilized peripheral blood.39,48,49 Transplantation of ZFN-edited HSPCs into NSG mice was used to evaluate the safety of the treatment, with readouts in the ability of the treated cells to engraft and differentiate in the same manner as unmodified cells and to give rise to gene modified CD4+ T-cell progeny.50 Evidence of efficacy was also demonstrated by the selective preservation of gene-modified cells during HIV infection and by the suppression of viremia to levels below the limit of detection in mouse plasma.39,51
The development of effective and clinically appropriate nuclease delivery methods for HSPCs has been a major challenge. Initial experiments using electroporation of plasmid DNA, although simple, ran into dose-limiting toxicities at higher amounts. Evaluation of Ad5/F35-mediated delivery into adult mobilized HSPCs revealed that this vector is less efficient in HSPCs than in CD4+ T cells, achieving only 5% CCR5 disruption, although frequencies could be increased by treatment with protein kinase C activators.48 Subsequently, we and others identified mRNA electroporation as an efficient and nontoxic method for ZFN introduction into HSPCs, achieving 30% to 50% CCR5 disruption with little toxicity.49,52,53 This method has now been adapted for large-scale clinical use in a recently initiated clinical trial (#NCT02500849) (Table 1).
Because HSPC gene therapy is often limited by difficulties in the ex vivo culture and expansion of HSPCs, there is also interest in the idea of modifying patient-specific induced pluripotent stem cells (iPSCs), which could then be reprogrammed into HSPCs.54 Clonal selection, differentiation, and expansion would allow 100% modification efficiencies to be achieved with homogeneity of the genome editing outcome while facilitating comprehensive testing for off-target effects. Several groups have reported making CCR5-disrupted CD34+ cells at small-scale from iPSCs using ZFNs, TALENs, and CRISPR/Cas9, and facilitated by HDR-mediated insertion of a selection cassette into CCR5.55,56 Further advances in reprogramming technologies may allow this to become a viable clinical alternative to modifying autologous HSPCs.
Future approaches to gene disruption
Beyond ZFNs, other nuclease platforms are also being developed to disrupt CCR5 in T cells and HSPCs, including TALENs,57 megaTALs,58 and CRISPR/Cas9.59,60 One potential advantage of CRISPR/Cas9 is that multiple guide RNAs can be used simultaneously to potentiate the magnitude of gene disruption or to induce specific deletions that are larger than the typical indels generated by NHEJ.59 However such multicomponent systems (requiring nuclease and guide RNAs) may introduce complexity into the clinical development process, and clinically suitable methods to deliver CRISPR/Cas9 to HSPCs in particular are still under development.61
Although early infecting HIV-1 strains use CCR5 as the coreceptor, CXCR4-tropic viruses emerge in nearly half of patients in the later stages of infection.23,62 Disruption of CXCR4 is therefore also being explored as a strategy for patients who harbor CXCR4-tropic HIV-1. However, because CXCR4 plays an essential role in hematopoietic stem cell homing and retention in the bone marrow, this approach is limited to T cells.63 Two reports have described the ability of CXCR4-targeted ZFNs to protect human CD4+ T cells in humanized mice infected with CXCR4-tropic HIV-164,65 while the simultaneous disruption of both CCR5 and CXCR4 by ZFNs conferred protection against both tropisms of virus.66
HDR-mediated gene editing and addition for anti-HIV therapies
More sophisticated application of genome editing will be achieved through exploiting HDR pathways. Compared with standard lentiviral vector approaches, HDR-mediated insertion of an anti-HIV gene offers the potential to provide greater control over the location, copy number, and expression profile of the added transgene. Those anti-HIV genes already evaluated in clinical trials remain candidates for this more precise method of gene addition.10-16 Moreover, by inserting such cassettes at a disrupted CCR5 locus, HIV resistance factors could be “stacked”67 and also provide protection to even mono-allelically modified cells. Beyond anti-HIV genes, genome editing technologies are also being developed to precisely insert HIV-specific chimeric antigen receptors into T cells and thereby exploit recent advances in immunotherapy to kill HIV-infected cells.58
The capability of genome engineering to make small changes to genes could also be exploited to engineer alleles of human genes that are associated with better virologic control. Two broad classes of host genes could be considered as candidates: restriction factors and dependency factors.
Cellular restriction factors are host genes with broad antiviral properties and are often interferon inducible.68 They are also characterized by high evolution rates, reflecting the ongoing battle between host factors and the pathogens they target. Restriction factors with activity against HIV-1 include TRIM5α, tetherin, and APOBEC3G, but the evolutionary adaptations of HIV-1 to its human host mean that the human versions no longer function against this virus. In contrast, orthologs from nonhuman primates frequently retain anti-HIV activity, and comparisons between human and primate genes have identified single or small numbers of amino acids that could be edited into the human genes to restore anti-HIV activity.69-71
In contrast, dependency factors72-74 are the group of cellular factors that are essential for HIV-1 replication and that are exploited by this viral parasite. Together with information from genome-wide association studies, which can identify genetic variants associated with greater HIV control,75 it may be possible to select candidate alleles from naturally occurring human variants that could also be engineered to inhibit HIV-1. The challenge will always be to find mutations that successfully prevent HIV from exploiting a host cell gene while not adversely impacting its function in noninfected cells.
Although these applications of genome editing suggest exciting new possibilities for HIV therapies, a major limitation to their development has been the low HDR editing efficiency that has been achieved in quiescent cell types such as mature T cells and HSPCs. One reason is that HDR is mainly active during the S and G2 phases of the cell cycle, but promoting cell cycling can adversely affect the function of quiescent cells. In addition, homology templates need to be present as DNA, which can cause cytotoxicity due to activation of DNA sensing pathways. Progress is now being made, as using integrase-defective lentiviral vectors as homology templates allowed gene editing rates of up to 5% at the IL2RG, CCR5, and AAVS1 loci in CD4+ T cells76 and at IL2RG in CD34+ HSCPs.53,76-78 More recently, we and others have described the use of adeno-associated virus (AAV) serotype 6 vectors in combination with electroporation of nuclease mRNA to achieved HDR-mediated genome editing. This method can be applied to both primary T cells and HSPCs, with little toxicity, and achieving editing rates of 8% to 60% at CCR5 alleles in primary T cells58,61 and 15% to 40% at the CCR5, IL2RG, HBB, and AAVS1 loci in mobilized blood HSPCs.49,58 Importantly, we also demonstrated that this approach edits the most primitive CD90+ cells in the bulk CD34+ population and can support serial transplantation of immune-deficient mice.49 Future methods to improve outcomes may come from manipulation of culture conditions to preserve gene-modified primitive HSPCs,53 or by the knocking-in of a selection marker, to allow further in vivo selection for successfully modified cells.79,80
Disrupting integrated HIV-1 proviral genome
A major limitation of ART and many anti-HIV gene therapies that target different stages of the viral life cycle is that, although these approaches can prevent spread of infection, they do not eliminate integrated HIV-1 genomes. Because an integrated genome is to all intents and purposes a gene in the cell that carries it, the potential exists to use the capabilities of engineered nucleases to disrupt these unwanted genetic passengers. Moreover, such applications could theoretically also target latent genomes and thereby reduce the size of the HIV-1 reservoir that persists despite ART.
The first use of engineered nucleases to excise integrated HIV-1 genomes was reported by Sarkar et al, who evolved a Cre recombinase (Tre) to recognize a sequence within the HIV-1 long terminal repeat that had 50% similarity to the natural Cre recognition site, LoxP.81 This approach takes advantage of the ability of Cre to remove the intervening DNA sequence between two flanking LoxP sites. More recently, the same group has evolved a more broadly acting enzyme, named universal Tre (uTre), that targets an long terminal repeat sequence conserved across multiple subtypes of HIV-1.82 More conventional anti-HIV nucleases have also been described based on homing endonuclease, ZFNs, TALENs, and CRISPR/Cas9, targeting different regions of the HIV-1 genome, and with a demonstrated ability to reduce HIV-1 content in various cell lines.83-88
Although these studies provide the initial proof of principle that gene disruption could be used to disrupt or excise an integrated HIV-1 genome, significant challenges will exist in the clinical translation of these approaches. No efficient method yet exists to enable in vivo delivery of nucleases to HIV target cells, and delivery to the subset of cells that comprise the latent reservoir will be especially problematic, due both to the location of these cells in inaccessible organ niches and the lack of identifying surface markers of viral infection. Alternatively, HIV-targeted nucleases could be delivered to bulk CD4+ T cells or HSPCs and be placed under inducible expression using an HIV-1 Tat-responsive promoter, as was reported in the case of the Tre recombinase.81 The recent reports of CD4-targeted lentiviral and AAV vectors suggest strategies toward targeted in vivo transduction of bulk CD4+ T cells.89,90
Summary
HIV/AIDS provides a uniquely suitable disease for the development of applications of genome editing. ZFNs are currently the most clinically advanced engineered nuclease system being used, with 7 phase 1/2 clinical trials evaluating CCR5 disruption in T cells and HSPCs. In the future, engineered nucleases could also be used to disrupt the alternative HIV-1 coreceptor, CXCR4, or to target integrated HIV genomes directly. Moreover, recent demonstrations that highly efficient gene editing or addition is now possible in T cells and HSPCs through the use of AAV vectors to deliver homology templates should allow even more sophisticated applications of genome editing against this important human pathogen.
Acknowledgments
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grant HL129902 and California Institute for Regenerative Medicine grant RT3-07848.
Authorship
Contribution: C.X.W. and P.M.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paula Cannon, Department of Molecular Microbiology & Immunology, Keck School of Medicine, University of Southern California, 2011 Zonal Ave, HMR413, Los Angeles, CA 90033; e-mail: pcannon@usc.edu.