Abstract
The red cell membrane skeleton is a pseudohexagonal meshwork of spectrin, actin, protein 4.1R, ankyrin, and actin-associated proteins that laminates the inner membrane surface and attaches to the overlying lipid bilayer via band 3–containing multiprotein complexes at the ankyrin- and actin-binding ends of spectrin. The membrane skeleton strengthens the lipid bilayer and endows the membrane with the durability and flexibility to survive in the circulation. In the 36 years since the first primitive model of the red cell skeleton was proposed, many additional proteins have been discovered, and their structures and interactions have been defined. However, almost nothing is known of the skeleton’s physiology, and myriad questions about its structure remain, including questions concerning the structure of spectrin in situ, the way spectrin and other proteins bind to actin, how the membrane is assembled, the dynamics of the skeleton when the membrane is deformed or perturbed by parasites, the role lipids play, and variations in membrane structure in unique regions like lipid rafts. This knowledge is important because the red cell membrane skeleton is the model for spectrin-based membrane skeletons in all cells, and because defects in the red cell membrane skeleton underlie multiple hemolytic anemias.
History
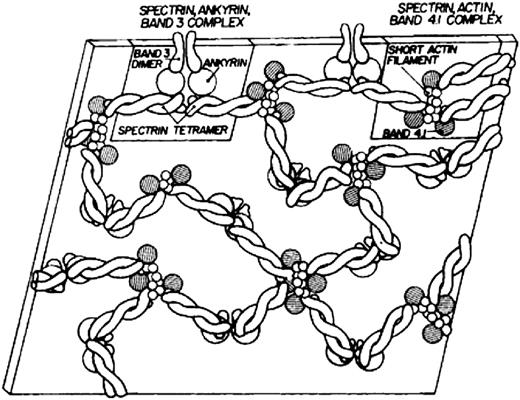
Modern knowledge of the red cell plasma membrane and its membrane skeleton began with Marchesi and Steers’s identification of spectrin in 1968.1 Prior to that year, almost the only thing known about membranes was that they contained a lipid bilayer.2 Indeed, there was a period in the 1960s when it was believed that red cell membranes contained only a single 22.5-kDa protein called “structural protein.”3 The fertile decade following the discovery of spectrin and the more or less coincident introduction of sodium dodecyl sulfate polyacrylamide gel electrophoresis as an analytic tool led to the isolation and characterization of several major red cell membrane proteins. The first simple model of the membrane skeleton appeared 36 years ago (Figure 1),4 and although it contained the core elements of the modern model, a great deal has been learned in the intervening decades (Figure 2 5 ). Hundreds of individuals have contributed to our knowledge of the red cell membrane skeleton, but a few deserve special recognition, including Peter Agre, David Anstee, G. Vann Bennett, Daniel Branton, Lesley Bruce, Jean Delaunay, David Discher, Velia Fowler, Patrick Gallagher, Walter Gratzer, Philip Low, Vincent Marchesi, Narla Mohandas, Jon Morrow, Jeri Palek, Luanne Peters, David Speicher, Ted Steck, and Michael Tanner.
The first model of the red cell membrane skeleton, published 36 years ago. Reprinted from Lux4 with permission.
The first model of the red cell membrane skeleton, published 36 years ago. Reprinted from Lux4 with permission.
Current model of the red cell membrane. Most of the known protein contacts are shown, but the relative positions of the proteins to each other within the various complexes are mostly not known. The major proteins are drawn roughly to scale, but the shapes are mostly imaginary. Approximately 40% of the band 3 molecules are tetramers in a complex with ankyrin and other integral proteins near the spectrin self-association site (Ankyrin complex). An approximately similar fraction of the band 3 molecules, probably dimers, are located near the spectrin-actin junction and bind to spectrin via protein 4.1R (4.1), protein 4.2 (4.2), and adducin (Actin junctional complex). As described later in the text, it is likely that these 2 complexes, with their associated proteins, are large enough that they sometimes contact each other. The remaining band 3 dimers float untethered within the lipid bilayer (Unbound band 3). The actin protofilament lies parallel to the membrane. The complexes of proteins associated with band 3 are not constant; that is, some proteins are present in much smaller numbers than others (eg, Kell, Kx, CD44, CD47, DARC/Duffy, LW, phosphofructokinase, and aldolase). The amounts of p55, adducin, and dematin are also insufficient to interact with all the spectrin/protein 4.1/actin complexes. As a consequence, the ankyrin and actin junctional complexes must vary in composition and mobility. For visual clarity, peroxiredoxin 2 (Prx2) is shown attached only to unbound band 3, but there is no evidence for this selectivity. CH, calponin homology; CH1 and CH2, actin binding domains of β-spectrin; EF, calcium ion–binding EF hand domain of α-spectrin; F-actin, filamentous actin; GEC, glycolytic enzyme complex (glyceraldehyde-3-phosphate dehydrogenase, phosphofructokinase, lactic dehydrogenase, pyruvate kinase, aldolase, and enolase); Glut1, glucose transporter 1; GPA, glycophorin A; GPB, glycophorin B; GPC/D, glycophorins C and D; LW, Landsteiner-Wiener; RhAG, Rh-associated glycoprotein. Professional illustration by Somersault18:24. Adapted from Korsgren et al5 with permission.
Current model of the red cell membrane. Most of the known protein contacts are shown, but the relative positions of the proteins to each other within the various complexes are mostly not known. The major proteins are drawn roughly to scale, but the shapes are mostly imaginary. Approximately 40% of the band 3 molecules are tetramers in a complex with ankyrin and other integral proteins near the spectrin self-association site (Ankyrin complex). An approximately similar fraction of the band 3 molecules, probably dimers, are located near the spectrin-actin junction and bind to spectrin via protein 4.1R (4.1), protein 4.2 (4.2), and adducin (Actin junctional complex). As described later in the text, it is likely that these 2 complexes, with their associated proteins, are large enough that they sometimes contact each other. The remaining band 3 dimers float untethered within the lipid bilayer (Unbound band 3). The actin protofilament lies parallel to the membrane. The complexes of proteins associated with band 3 are not constant; that is, some proteins are present in much smaller numbers than others (eg, Kell, Kx, CD44, CD47, DARC/Duffy, LW, phosphofructokinase, and aldolase). The amounts of p55, adducin, and dematin are also insufficient to interact with all the spectrin/protein 4.1/actin complexes. As a consequence, the ankyrin and actin junctional complexes must vary in composition and mobility. For visual clarity, peroxiredoxin 2 (Prx2) is shown attached only to unbound band 3, but there is no evidence for this selectivity. CH, calponin homology; CH1 and CH2, actin binding domains of β-spectrin; EF, calcium ion–binding EF hand domain of α-spectrin; F-actin, filamentous actin; GEC, glycolytic enzyme complex (glyceraldehyde-3-phosphate dehydrogenase, phosphofructokinase, lactic dehydrogenase, pyruvate kinase, aldolase, and enolase); Glut1, glucose transporter 1; GPA, glycophorin A; GPB, glycophorin B; GPC/D, glycophorins C and D; LW, Landsteiner-Wiener; RhAG, Rh-associated glycoprotein. Professional illustration by Somersault18:24. Adapted from Korsgren et al5 with permission.
This review focuses on our current understanding of the anatomy of the membrane skeleton, particularly its spectrin-actin core, and on some of the key unanswered questions. Because of space limitations, many important aspects of the red cell membrane are not discussed. These include integral membrane proteins not attached to the skeleton, the lipid bilayer, membrane biogenesis, posttranslational modifications of the membrane, red cell aging, and mouse and human membrane diseases. For interested readers, a few other recent reviews are recommended.6-10
The red cell membrane skeleton
The red cell membrane contains ∼20 major proteins and at least 850 minor ones.11 Some of the important ones are described in Table 1. The integral membrane proteins are organized into macromolecular complexes centered on band 3, the anion-exchange channel. Most of the peripheral membrane proteins form the membrane skeleton, a protein meshwork 40- to 90-nm thick that laminates the inner membrane surface. The skeleton is composed principally of spectrin, actin and its associated proteins (tropomyosin, tropomodulin, adducin, and dematin), protein 4.1R, and ankyrin.
Some important erythrocyte membrane proteins
| Protein . | Molecular mass, kDa* . | Monomers per cell† × 10−3 . | Oligomeric state . | Oligomers in ankyrin complex . | Oligomers in actin junction complex . | Functions in mature erythrocyte membranes . | |
|---|---|---|---|---|---|---|---|
| Complex . | Unit cell‡ . | Complex and unit cell‡ . | |||||
| β-Actin | 41.6 | 500 | Oligomer (∼14) | 1 | Forms ∼37-nm long filaments that serve as a molecular junction in the membrane skeleton. Binds spectrin with the aid of protein 4.1R. Binds tropomyosin, tropomodulin, adducin, dematin, and chorein. | ||
| α-Adducin | 81.0 | 30 | Heterodimer/ tetramer | 1 (dimer) | Caps the fast-growing (barbed) end of actin and recruits spectrin to nearby sites on actin. Links actin junction to band 3, Glut1, and maybe stomatin. Binds chorein. | ||
| β-Adducin | 80.7 | 30 | |||||
| Aldolase A | 39.4 | 20 | Tetramer | <1 | <1 | <1 | Forms glycolytic complex on band 3 with PFK, enolase, G3PD, PK, and LDH. |
| Ankyrin-1§ | 206.0 | 124 ± 11 | Monomer | 1 | 3 | Attaches β-spectrin and the membrane skeleton to band 3 and to protein 4.2 and RhAG in the ankyrin-linked band 3 complex. | |
| Band 3 (SLC4A1) | 101.8 | 1200 | Dimer or tetramer | 2 (dimer) | 6 (dimer) | 1-6 (dimer) | Anion-exchange channel (especially HCO3− and Cl−) with the aid of CAII and CAIV. Forms membrane complexes with GPA, GPB, the Rh complex, Glut1, stomatin, and other proteins that are linked to spectrin via ankyrin-1 and protein 4.2, and linked to the actin junctional complex via protein 4.1R, protein 4.2, p55, adducin, and Glut1. Binds peroxiredoxin. Forms a glycolytic metabolon with G3PD, PFK, PGK, aldolase, LDH, and PK. Binds deoxyhemoglobin. Carries Ii and many other blood group antigens. |
| CAII | 29.1 | 100 | Unknown | Attaches to the outside (CAIV) and inside (CAII) surfaces of band 3, the HCO3− channel. Catalyzes interconversion of CO2 and HCO3−, which aids in HCO3− transport and CO2 elimination. | |||
| CAIV | 33.1 | Unknown | Unknown | ||||
| CD44 | 37.1 | 6-10 | Unknown | <1 | Binds to protein 4.1R and maybe to ankyrin. Multifunctional protein in nonerythroid cells. Function in the red cell is unknown. | ||
| CD47 | 33.3 | 17 | Monomer | <1 | <1 | Binds to protein 4.2 and to the Rh complex (RhAG/RhD/RhCE) within the ankyrin-linked band 3 complex. Inhibits phagocytosis. | |
| DARC/Duffy | 35.6 | 6-13 | Unknown | <1 | Chemokine receptor. Carries the Duffy (Fy) blood group antigen. Receptor for Plasmodium vivax. Function in normal red cell unknown. | ||
| Dematin | 43.1/45.5 | 130 | Monomer or trimer? | 3 (if monomer) 1 (if trimer) | Binds and bundles actin filaments. Facilitates spectrin-actin binding. Helps attach actin to the membrane via Glut1. | ||
| G3PD | 35.9 | 500 | Tetramer | 1 | 3 | 1-3 | Forms glycolytic metabolon on band 3 with aldolase, PFK, enolase, PK, and LDH. |
| Glut1 | 54.1 | 200-700 | Dimer and tetramer | ? | ? | 1-6 (dimer) | Transports glucose. Transports l-dehydroascorbic acid when bound to stomatin. Links actin to the membrane via interactions with dematin, adducin, and band 3. |
| GPA | 14.3 | 1000 | Homo or heterodimer | 2 | 6 | 1-6 | Binds band 3 in the ankyrin-linked band 3 complex, and maybe to RhAG. May facilitate band 3 transfer to the membrane. Carries MNSsU blood group antigens and much of the red cell’s negative surface charge. |
| GPB | 7.7 | 170-250 | |||||
| GPC | 13.8 | 143 | Monomer? Homo or heterodimer? | 6 | Links actin junctional complex to membrane via interactions with protein 4.1R, p55, and band 3. Carries Gerbich blood group antigens. | ||
| GPD | 11.5 | 82 | |||||
| Kell | 82.8 | 3-18 | Heterodimer with Kx | <1 | The Kx/Kell complex may reside in the actin-linked band 3 complex. Kell carries the Kell blood group antigen. Kx probably has an undiscovered transport function. | ||
| Kx | 50.9 | Unknown | Heterodimer with Kell | <1 | |||
| Lu/BCAM | 64.3 | 1.5-8 | Unknown | Adhesion molecule that carries the Lu blood group antigen. Binds to α-spectrin. | |||
| LW glycoprotein/ ICAM4 | 27.0 | 3-5 | Monomer | <1 | <1 | ICAM that carries the LW blood group antigen. Interacts with RhAG/RhD/RhCE and probably resides in the ankyrin-linked band 3 complex. | |
| p55/Palmitoylated membrane protein 1 | 52.3 | 80 | Dimer? | 1 | Links actin junctional complex to membrane via interactions with some protein 4.1R and GPC/D molecules. Heavily palmitoylated. May contribute to organization of lipid rafts. | ||
| Peroxiredoxin 2 | 21.8 | 200 | Dimer or decamer | ? | ? | ? | Abundant antioxidant protein that is partially linked to band 3 on the membrane. Also binds stomatin and activates the calcium ion–dependent potassium ion (Gardos) channel. |
| PFK (liver) | 85.0 | 6 | Heterotetramer | <1 | <1 | <1 | Forms glycolytic metabolon on band 3 with aldolase, G3PD, enolase, PK, and LDH. |
| PFK (muscle) | 85.2 | ||||||
| Protein 4.1R | 66.4 | ∼240 | Monomer | 6 | Binds to both β-spectrin and actin and greatly strengthens the spectrin-actin interaction. Links actin to the membrane via interactions with GPC/D, p55, and band 3. Competes with ankyrin for binding to band 3. | ||
| Protein 4.2 | 79.8/76.9 | ∼280 | Monomer? | 2 | 6 | 1 | Binds to band 3, ankyrin, and CD47 in the ankyrin-linked band 3 complex. Also binds to the EF hand domain of α-spectrin. |
| RhAG | 44.2 | 100-200 | Heterotrimer of RhAG, RhD, and RhCE? | 1 | 3 | The RhAG/RhD/RhCE trimer binds to CD47, LW, ankyrin, CD47, and GPB in the ankyrin-linked band 3 complex. RhAG transports NH3/NH4+ and maybe CO2. | |
| RhCE | 45.4 | 100-200 | |||||
| RhD | 45.1 | 100-200 | |||||
| α-Spectrin | 280.0 | 242 | Heterodimer/ tetramer/oligomer | 2 (dimer) | 6 (dimer) | 6 (dimer) | Cross-links actin filaments into a hexagonal lattice at the tail end with the help of protein 4.1R, adducin, and dematin. Binds protein 4.2 via the α-spectrin EF hand domain. Self-associates into tetramers and oligomers and binds to ankyrin-1 and Lu/BCAM at the head end. |
| β-Spectrin | 246.4 | 242 | |||||
| Stomatin | 31.7 | 200-400? | Oligomer (9-12)? | ? | ? | 1 | Binds to Glut1 and converts it into an ascorbic acid transporter. Also binds band 3, adducin, and peroxiredoxin. Located in lipid rafts. |
| Tropomodulin 1 | 40.6 | 30 | Monomer | 1 | Caps slow-growing (pointed) end of actin. Binds TM, which strengthens capping. | ||
| Tropomyosin 1 (α-TM) | 32.9 | 140 | Homodimer | 2 | Binds to and stabilizes actin filaments in a magnesium ion–dependent manner. May help specify actin filament lengths in the red cell. | ||
| Tropomyosin 3 (γ-TM) | 32.9 | ||||||
| Protein . | Molecular mass, kDa* . | Monomers per cell† × 10−3 . | Oligomeric state . | Oligomers in ankyrin complex . | Oligomers in actin junction complex . | Functions in mature erythrocyte membranes . | |
|---|---|---|---|---|---|---|---|
| Complex . | Unit cell‡ . | Complex and unit cell‡ . | |||||
| β-Actin | 41.6 | 500 | Oligomer (∼14) | 1 | Forms ∼37-nm long filaments that serve as a molecular junction in the membrane skeleton. Binds spectrin with the aid of protein 4.1R. Binds tropomyosin, tropomodulin, adducin, dematin, and chorein. | ||
| α-Adducin | 81.0 | 30 | Heterodimer/ tetramer | 1 (dimer) | Caps the fast-growing (barbed) end of actin and recruits spectrin to nearby sites on actin. Links actin junction to band 3, Glut1, and maybe stomatin. Binds chorein. | ||
| β-Adducin | 80.7 | 30 | |||||
| Aldolase A | 39.4 | 20 | Tetramer | <1 | <1 | <1 | Forms glycolytic complex on band 3 with PFK, enolase, G3PD, PK, and LDH. |
| Ankyrin-1§ | 206.0 | 124 ± 11 | Monomer | 1 | 3 | Attaches β-spectrin and the membrane skeleton to band 3 and to protein 4.2 and RhAG in the ankyrin-linked band 3 complex. | |
| Band 3 (SLC4A1) | 101.8 | 1200 | Dimer or tetramer | 2 (dimer) | 6 (dimer) | 1-6 (dimer) | Anion-exchange channel (especially HCO3− and Cl−) with the aid of CAII and CAIV. Forms membrane complexes with GPA, GPB, the Rh complex, Glut1, stomatin, and other proteins that are linked to spectrin via ankyrin-1 and protein 4.2, and linked to the actin junctional complex via protein 4.1R, protein 4.2, p55, adducin, and Glut1. Binds peroxiredoxin. Forms a glycolytic metabolon with G3PD, PFK, PGK, aldolase, LDH, and PK. Binds deoxyhemoglobin. Carries Ii and many other blood group antigens. |
| CAII | 29.1 | 100 | Unknown | Attaches to the outside (CAIV) and inside (CAII) surfaces of band 3, the HCO3− channel. Catalyzes interconversion of CO2 and HCO3−, which aids in HCO3− transport and CO2 elimination. | |||
| CAIV | 33.1 | Unknown | Unknown | ||||
| CD44 | 37.1 | 6-10 | Unknown | <1 | Binds to protein 4.1R and maybe to ankyrin. Multifunctional protein in nonerythroid cells. Function in the red cell is unknown. | ||
| CD47 | 33.3 | 17 | Monomer | <1 | <1 | Binds to protein 4.2 and to the Rh complex (RhAG/RhD/RhCE) within the ankyrin-linked band 3 complex. Inhibits phagocytosis. | |
| DARC/Duffy | 35.6 | 6-13 | Unknown | <1 | Chemokine receptor. Carries the Duffy (Fy) blood group antigen. Receptor for Plasmodium vivax. Function in normal red cell unknown. | ||
| Dematin | 43.1/45.5 | 130 | Monomer or trimer? | 3 (if monomer) 1 (if trimer) | Binds and bundles actin filaments. Facilitates spectrin-actin binding. Helps attach actin to the membrane via Glut1. | ||
| G3PD | 35.9 | 500 | Tetramer | 1 | 3 | 1-3 | Forms glycolytic metabolon on band 3 with aldolase, PFK, enolase, PK, and LDH. |
| Glut1 | 54.1 | 200-700 | Dimer and tetramer | ? | ? | 1-6 (dimer) | Transports glucose. Transports l-dehydroascorbic acid when bound to stomatin. Links actin to the membrane via interactions with dematin, adducin, and band 3. |
| GPA | 14.3 | 1000 | Homo or heterodimer | 2 | 6 | 1-6 | Binds band 3 in the ankyrin-linked band 3 complex, and maybe to RhAG. May facilitate band 3 transfer to the membrane. Carries MNSsU blood group antigens and much of the red cell’s negative surface charge. |
| GPB | 7.7 | 170-250 | |||||
| GPC | 13.8 | 143 | Monomer? Homo or heterodimer? | 6 | Links actin junctional complex to membrane via interactions with protein 4.1R, p55, and band 3. Carries Gerbich blood group antigens. | ||
| GPD | 11.5 | 82 | |||||
| Kell | 82.8 | 3-18 | Heterodimer with Kx | <1 | The Kx/Kell complex may reside in the actin-linked band 3 complex. Kell carries the Kell blood group antigen. Kx probably has an undiscovered transport function. | ||
| Kx | 50.9 | Unknown | Heterodimer with Kell | <1 | |||
| Lu/BCAM | 64.3 | 1.5-8 | Unknown | Adhesion molecule that carries the Lu blood group antigen. Binds to α-spectrin. | |||
| LW glycoprotein/ ICAM4 | 27.0 | 3-5 | Monomer | <1 | <1 | ICAM that carries the LW blood group antigen. Interacts with RhAG/RhD/RhCE and probably resides in the ankyrin-linked band 3 complex. | |
| p55/Palmitoylated membrane protein 1 | 52.3 | 80 | Dimer? | 1 | Links actin junctional complex to membrane via interactions with some protein 4.1R and GPC/D molecules. Heavily palmitoylated. May contribute to organization of lipid rafts. | ||
| Peroxiredoxin 2 | 21.8 | 200 | Dimer or decamer | ? | ? | ? | Abundant antioxidant protein that is partially linked to band 3 on the membrane. Also binds stomatin and activates the calcium ion–dependent potassium ion (Gardos) channel. |
| PFK (liver) | 85.0 | 6 | Heterotetramer | <1 | <1 | <1 | Forms glycolytic metabolon on band 3 with aldolase, G3PD, enolase, PK, and LDH. |
| PFK (muscle) | 85.2 | ||||||
| Protein 4.1R | 66.4 | ∼240 | Monomer | 6 | Binds to both β-spectrin and actin and greatly strengthens the spectrin-actin interaction. Links actin to the membrane via interactions with GPC/D, p55, and band 3. Competes with ankyrin for binding to band 3. | ||
| Protein 4.2 | 79.8/76.9 | ∼280 | Monomer? | 2 | 6 | 1 | Binds to band 3, ankyrin, and CD47 in the ankyrin-linked band 3 complex. Also binds to the EF hand domain of α-spectrin. |
| RhAG | 44.2 | 100-200 | Heterotrimer of RhAG, RhD, and RhCE? | 1 | 3 | The RhAG/RhD/RhCE trimer binds to CD47, LW, ankyrin, CD47, and GPB in the ankyrin-linked band 3 complex. RhAG transports NH3/NH4+ and maybe CO2. | |
| RhCE | 45.4 | 100-200 | |||||
| RhD | 45.1 | 100-200 | |||||
| α-Spectrin | 280.0 | 242 | Heterodimer/ tetramer/oligomer | 2 (dimer) | 6 (dimer) | 6 (dimer) | Cross-links actin filaments into a hexagonal lattice at the tail end with the help of protein 4.1R, adducin, and dematin. Binds protein 4.2 via the α-spectrin EF hand domain. Self-associates into tetramers and oligomers and binds to ankyrin-1 and Lu/BCAM at the head end. |
| β-Spectrin | 246.4 | 242 | |||||
| Stomatin | 31.7 | 200-400? | Oligomer (9-12)? | ? | ? | 1 | Binds to Glut1 and converts it into an ascorbic acid transporter. Also binds band 3, adducin, and peroxiredoxin. Located in lipid rafts. |
| Tropomodulin 1 | 40.6 | 30 | Monomer | 1 | Caps slow-growing (pointed) end of actin. Binds TM, which strengthens capping. | ||
| Tropomyosin 1 (α-TM) | 32.9 | 140 | Homodimer | 2 | Binds to and stabilizes actin filaments in a magnesium ion–dependent manner. May help specify actin filament lengths in the red cell. | ||
| Tropomyosin 3 (γ-TM) | 32.9 | ||||||
Ankyrin-1, erythrocyte ankyrin; BCAM, basal cell adhesion molecule; CAII, carbonic anhydrase II; CAIV, carbonic anhydrase IV; Cl−, chloride ion; CO2, carbon dioxide; G3PD, glyceraldehyde-3-phosphate dehydrogenase; HCO3−, bicarbonate; ICAM4, intercellular adhesion molecule 4; LDH, lactic dehydrogenase; Lu, Lutheran; NH3/NH4+, ammonia/ammonium ion; PFK, phosphofructokinase; PGK, phosphoglycerate kinase; PK, pyruvate kinase; TM, tropomyosin.
Size of the mature protein chain without any signal peptide, propeptides, or initiator methionine, but not including posttranslational modifications.
References for the numbers of copies of each protein are listed in the footnotes of Table 2 found in Lux6 . It must be emphasized that the numbers of many red cell membrane proteins are roughly estimated but not accurately measured, which affects the estimates of the composition of the ankyrin complex and actin junctional complex.
The unit cell is defined in Figure 8.
Data shown are for full-length ankyrin (band 2.1), the major isoform, except for the number of copies per cell, which includes the smaller isoforms: bands 2.2, 2.3, and 2.6.
Spectrin
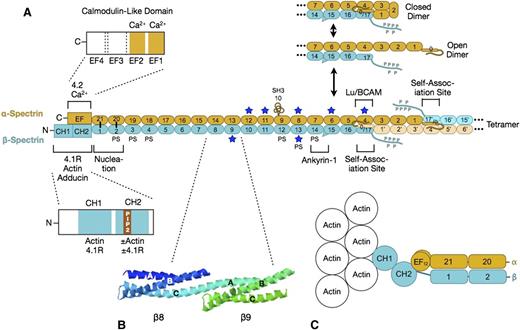
Erythrocyte spectrin is a long, flexible, wormlike protein composed of 2 parallel chains (α- and β-spectrin) oriented in opposite directions. Each chain contains multiple spectrin-type repeats, with specialized functional domains at the “head” end for spectrin dimer-tetramer association and for erythrocyte ankyrin (also known as ankyrin-1 or ankyrin-R) binding, and domains at the “tail” end for binding to protein 4.1R, protein 4.2, short filaments of actin, and other proteins. The details of spectrin structure are shown in Figure 3.12-26 On average, 6 spectrins bind per actin filament, leading to a pseudohexagonal arrangement.27
Spectrin. (A) Organization of erythrocyte spectrin and the dimer-tetramer equilibrium. Spectrin is a long, flexible, wormlike protein composed of 2 chains (α- and β-spectrin). Each chain contains a tandem array of ∼5.0-nm, ∼106-amino acid, triple-helical spectrin-type repeats, with specialized functional domains for self-association and ankyrin-1 binding at the head end, and for binding to actin, protein 4.1R, and other associated proteins at the tail end. Each spectrin repeat is formed by 3 α-helices (A, B, and C) with short connecting loops that are folded like a flattened Z into a triple-helical bundle.12 α-Spectrin contains 21 numbered repeats (α1-α21), plus a partial repeat (α0) at the N-terminus that contains a single C-helix. One of the 21 is really an src homology 3 (SH3) domain (α10) but is numbered as a repeat by convention. β-Spectrin contains 16 true repeats (β1-β16) plus a partial repeat (β17) at the C-terminus that contains just the A and B helices. Note that for the spectrin αβ dimer to convert to the α2β2 tetramer, it must first cleave the internal linkage between the partial spectrin repeats α0 and β17 and then unfold (open dimer).13 This is the rate-limiting step in the dimer-tetramer equilibrium. Dimer-tetramer self-association occurs at the head end of the spectrin dimer where the adhesion protein Lutheran/basal cell adhesion molecule (Lu/BCAM) also attaches.14 Ankyrin-1 binds nearby to spectrin repeats β14-β15.15 These 2 reactions cooperate: ankyrin-binding favors spectrin tetramer formation, and vice versa.16 The isolated α- and β-spectrin chains join to form spectrin heterodimers at a nucleation site near the tail end of spectrin (repeats α-21 pairs with β-1, and α-20 with β-2) and then zip together in a cooperative manner.17 Actin and protein 4.1R bind to CH domains at the N-terminal end of β-spectrin, just beyond the nucleation site.18 Binding to the CH2 domain is activated by phosphatidylinositol-4,5-bisphosphate (PIP2).18 Adducin binds in the same region.19 Protein 4.2 and calcium ion (Ca2+) bind to a neighboring EF hand domain on α-spectrin.5 Both the EF hand and CH domains are needed for full actin binding.20,21 PS denotes spectrin repeats that bind phosphatidyl serine.22 Blue stars mark repeats that are relatively unstable at physiological temperatures.23 (B) Structure of spectrin repeats β8 and β9 (PDB 1S3524 ). Note that each repeat is formed by 3 α-helices (A, B, and C) in a Z configuration. Note also that helix C in β8 and helix A in β9 form a continuous α-helix that spans the junction between the repeats. (C) Hypothetical model of the tail end of spectrin based on recent structures of α-actinin.25,26 Note that the first actin-binding domain (CH1) binds to F-actin in an extended (open) conformation.26 The intimate relationship between the EF hand and CH domains and the recent evidence that the EF hand domain is required for optimal spectrin-actin binding20,21 suggest that the EF hand domains regulate actin binding.
Spectrin. (A) Organization of erythrocyte spectrin and the dimer-tetramer equilibrium. Spectrin is a long, flexible, wormlike protein composed of 2 chains (α- and β-spectrin). Each chain contains a tandem array of ∼5.0-nm, ∼106-amino acid, triple-helical spectrin-type repeats, with specialized functional domains for self-association and ankyrin-1 binding at the head end, and for binding to actin, protein 4.1R, and other associated proteins at the tail end. Each spectrin repeat is formed by 3 α-helices (A, B, and C) with short connecting loops that are folded like a flattened Z into a triple-helical bundle.12 α-Spectrin contains 21 numbered repeats (α1-α21), plus a partial repeat (α0) at the N-terminus that contains a single C-helix. One of the 21 is really an src homology 3 (SH3) domain (α10) but is numbered as a repeat by convention. β-Spectrin contains 16 true repeats (β1-β16) plus a partial repeat (β17) at the C-terminus that contains just the A and B helices. Note that for the spectrin αβ dimer to convert to the α2β2 tetramer, it must first cleave the internal linkage between the partial spectrin repeats α0 and β17 and then unfold (open dimer).13 This is the rate-limiting step in the dimer-tetramer equilibrium. Dimer-tetramer self-association occurs at the head end of the spectrin dimer where the adhesion protein Lutheran/basal cell adhesion molecule (Lu/BCAM) also attaches.14 Ankyrin-1 binds nearby to spectrin repeats β14-β15.15 These 2 reactions cooperate: ankyrin-binding favors spectrin tetramer formation, and vice versa.16 The isolated α- and β-spectrin chains join to form spectrin heterodimers at a nucleation site near the tail end of spectrin (repeats α-21 pairs with β-1, and α-20 with β-2) and then zip together in a cooperative manner.17 Actin and protein 4.1R bind to CH domains at the N-terminal end of β-spectrin, just beyond the nucleation site.18 Binding to the CH2 domain is activated by phosphatidylinositol-4,5-bisphosphate (PIP2).18 Adducin binds in the same region.19 Protein 4.2 and calcium ion (Ca2+) bind to a neighboring EF hand domain on α-spectrin.5 Both the EF hand and CH domains are needed for full actin binding.20,21 PS denotes spectrin repeats that bind phosphatidyl serine.22 Blue stars mark repeats that are relatively unstable at physiological temperatures.23 (B) Structure of spectrin repeats β8 and β9 (PDB 1S3524 ). Note that each repeat is formed by 3 α-helices (A, B, and C) in a Z configuration. Note also that helix C in β8 and helix A in β9 form a continuous α-helix that spans the junction between the repeats. (C) Hypothetical model of the tail end of spectrin based on recent structures of α-actinin.25,26 Note that the first actin-binding domain (CH1) binds to F-actin in an extended (open) conformation.26 The intimate relationship between the EF hand and CH domains and the recent evidence that the EF hand domain is required for optimal spectrin-actin binding20,21 suggest that the EF hand domains regulate actin binding.
Isolated α- and β-spectrin chains bind to each other electrostatically at a pair of nucleation sites formed by repeats near the spectrin tail (Figure 3A). After this initial connection, the rest of the α and β chains zip together, coiling around each other approximately every 428 repeats. The side-to-side interactions beyond the nucleation site are relatively weak,29 allowing the 2 chains to slide past each other when the spectrin molecule flexes and extends during membrane deformation.
Spectrin α and β chains associate with each other at the head end. Basically, the complementary partial repeats (α0 and β17) at the end of each chain join to form a complete 3-helix repeat: α0/β17 (Figure 3A). In spectrin αβ dimers, the longer α chain folds back on itself, so the α0/β17 repeat interacts with repeat α4 and the α1 and α3 repeats contact each other (Figure 3A).13 In the α2β2-heterotetramers, there are 3 such side-to-side interactions between the longer α chains (Figure 3C). These lateral interactions, though weak, contribute to self-association. Higher oligomers (hexamers, octamers, and so forth) form by the same mechanism and can also be seen on the membrane.27,30,31 Even though nearly 95% of the spectrin is in the tetramer or oligomer forms,32 spectrin self-association is uniquely weak in the red cell.33 Tetramers dissociate and reform under physiological conditions, and this is greatly accentuated when the membrane is distorted by shear forces.34 This mechanism may be an evolutionary accommodation to permit the enormous distortions that the red cell undergoes in parts of the microvasculature.
Spectrin is a highly flexible molecule, but it is not certain how it achieves its flexibility because crystal structures of tandem repeats show a continuous helix across the junction between repeats (Figure 3B). There is evidence that the helical linker may bend in certain directions at the junction35 and that helices within some repeats may “melt” and rearrange, shortening the repeat.35 In addition, some of the repeats are unstable at physiological temperatures (starred repeats in Figure 3A)23 and may partially unfold when red cells deform, which would contribute to flexibility.36
The actin protofilament and its associated proteins
Actin.
Red cells contain short, double-helical filaments of nonmuscle or β-actin, termed “protofilaments” (Figure 2). These lie roughly parallel to the membrane plane (±20°) and are randomly oriented.37 There are ∼30 000 to 40 000 protofilaments per red cell, with 6 to 8 actin monomers in each of the 2 strands.
Not much is known about how these unique actin filaments form, but some hints are emerging from defects in actin assembly. Mice that lack both Rac1 and Rac2 GTPases,38 or lack Hem-1, a member of the Wiskott-Aldrich syndrome verprolin-homologous (WAVE) protein complex that regulates F-actin polymerization,39 develop hemolytic anemias with misshaped red cells and clumped or irregular skeletons. This finding suggests that Rac GTPases and Hem-1 help organize or regulate the red cell membrane skeleton, acting through pathways that stimulate actin polymerization in many cell types. It is unclear whether such regulation occurs dynamically in the mature red cell or just during erythropoiesis.
Tropomyosin.
Erythrocyte tropomyosin (TM) is a long rodlike dimer of α- and γ-TM isoforms (hTM5b and hTM5, respectively).10 One TM dimer binds to each of the 2 strands in the actin protofilament (Figure 2). Binding is magnesium ion dependent. The red cell TM isoforms also bind to tropomodulin (see below). Red cell membranes that lack TM are markedly more fragile than normal,40 and only endogenous TM, not the longer muscle isoforms, can restore stability. Red cell TM is just long enough (∼34 nm) to cover the estimated 7 actin monomers in 1 strand of the F-actin protofilament, which supports the idea that TM both serves as a molecular ruler (along with actin-capping proteins) in assembling the filament41 and strengthens the filament after assembly.
In yeast, formins (a family of proteins that nucleate actin filaments) direct which TM isoform associates with an actin filament.42 It will be interesting to see whether that is also true in erythroid precursors and whether formins play a role in protofilament formation.
Tropomodulin 1.
Tropomodulin 1 (TMod1) has 2 functions: it caps the pointed or slow-growing end of actin filaments (Figure 2) and binds TM, which greatly strengthens actin capping.43 There are 30 000 copies per red cell, or 1 per actin protofilament. One TMod1 binds to both actin strands and both TM dimers in the actin protofilament.43 Mice that lack TMod1 in their red cells have variable actin protofilament lengths and irregular holes in the membrane skeleton.44 These mice also have increased tropomodulin 3 in their red cells, which may partially compensate. Tropomodulin 3 is a protein that plays multiple roles in definitive erythropoiesis, erythroblast binding to so-called nurse macrophages, and enucleation,45 but is not normally present in mature red cells.
Protein 4.1R.
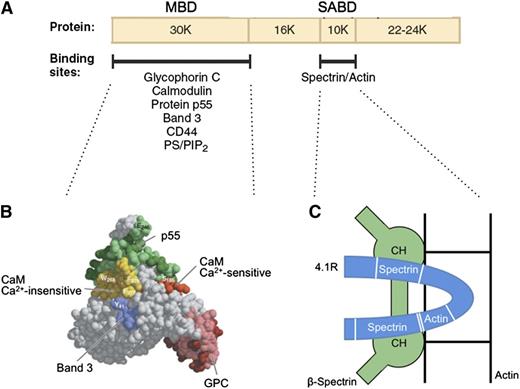
Protein 4.1R promotes spectrin-actin binding and helps attach the membrane skeleton to the membrane. The first of these functions is especially important. Under physiological conditions, erythrocyte spectrin binds very weakly to F-actin. Protein 4.1R binds to both spectrin and actin and generates a high-affinity ternary complex.46 The cofactor activity is contained in a 10-kDa spectrin-actin binding domain (SABD) in the middle of the molecule47 (Figure 448,49 ). The actin-binding site in the SABD is straddled by a 2-part spectrin-binding site in a single linear peptide (Figure 4C).49 Red cells that lack protein 4.1R are very fragile, but their membrane strength is normalized by the SABD peptide.50 It is unfortunate that the 3-dimensional structure of the SABD within protein 4.1R is unknown. Solving the complete structure of this important protein is a major need in the field.
Protein 4.1R. (A) Domain map of protein 4.1R and the location of erythrocyte binding partners. (B) Structure of the protein 4.1R membrane binding domain (MBD).48 Subdomains where specific proteins bind are colored and labeled. (C) Schematic representation of the protein 4.1R SABD (blue). Two spectrin binding regions straddle an actin binding peptide.49 In this hypothetical model, each of the spectrin binding regions is assumed to interact with a different one of the 2 CH domains of β-spectrin, and both CH domains are assumed to interact with actin.49 Neither of these assumptions has been proved. Ca2+, calcium ion; CaM, calmodulin; PS/PIP2, phosphatidyl serine/phosphatidylinositol-4,5-bisphosphate. Panel B adapted from Han et al48 with permission.
Protein 4.1R. (A) Domain map of protein 4.1R and the location of erythrocyte binding partners. (B) Structure of the protein 4.1R membrane binding domain (MBD).48 Subdomains where specific proteins bind are colored and labeled. (C) Schematic representation of the protein 4.1R SABD (blue). Two spectrin binding regions straddle an actin binding peptide.49 In this hypothetical model, each of the spectrin binding regions is assumed to interact with a different one of the 2 CH domains of β-spectrin, and both CH domains are assumed to interact with actin.49 Neither of these assumptions has been proved. Ca2+, calcium ion; CaM, calmodulin; PS/PIP2, phosphatidyl serine/phosphatidylinositol-4,5-bisphosphate. Panel B adapted from Han et al48 with permission.
Adducin.
Adducin is a complex multifunctional protein containing an α subunit, and either a β subunit or, less often, a related γ subunit.51 The adducin subunits contain a globular head domain and an extended, flexible tail.51 The predominant form is probably the αβ heterodimer.52 There are 30 000 copies of the dimer per red cell.
As suggested by the number of copies, adducin caps the fast-growing or “barbed” end of the actin protofilaments in red cells (Figure 2).52 It also recruits spectrin to neighboring sites on actin,53 which enhances the capping ∼10-fold.50 These interrelated functions require a domain at the end of the adducin tail.50 Both functions are regulated by calmodulin and by several protein kinases.54 The effects of phosphorylation are complex, and it is not clear whether they are physiologically important.55 Indeed, the importance of adducin itself is uncertain, because mice that lack the protein have only mild hemolysis and spherocytosis.56
Adducin also helps attach the membrane skeleton to the lipid bilayer through interactions with the anion-exchange channel (band 3)57 and glucose transporter type 1 (Figure 2).58 The band 3 binding site is located in the tails of α- and β-adducin.57 Whether adducin binds dimers or tetramers of band 3 and whether it binds 1 or 2 of each is unknown.
Dematin.
Dematin is an actin-binding protein with 2 actin binding sites: one in its headpiece and one in its tail.59 The recombinant protein is a monomer,59 but the native protein may be a trimer. Dematin was first identified by its ability to bundle actin filaments into cables,60 but in mature red cells, where there are no such cables, dematin has 2 functions. First, unmodified dematin binds to spectrin and enhances spectrin-actin binding.61 This function is lost when dematin is phosphorylated by protein kinase A.59,61 Second, dematin binds to the glucose transporter58 and helps attach actin to the membrane.
How does spectrin bind to the actin protofilament?
Surprisingly little is known about this central question. Actin and protein 4.1 bind to 2 tandem CH domains at the N-terminal (or tail) end of β-spectrin (Figure 3A). The CH1 domain is constitutively active but the CH2 domain is incipient.18 It is unmasked by removal of a blocking helix or by phosphatidylinositol-4,5-bisphosphate, a lipid involved in many signaling pathways.18 It is not known how, or even if, phosphatidylinositol-4,5-bisphosphate regulates the CH2 domain in the native red cell.
The neighboring calmodulin-like EF hand domain of α-spectrin is also important. Deletion of the last 13 amino acids of the domain in a minispectrin or in the severely hemolytic sph1J mouse nearly ablates actin binding.21
The relationships among the EF hand domain, the CH domains, and F-actin are not known in spectrin but probably resemble those in better studied spectrin family members such as α-actinin,25,26,62 utrophin,63 dystrophin, fimbrin, and plectin (Figure 3C). The problem is that these proteins bind actin in different ways. Sometimes the CH domains are in contact with each other (closed position), and sometimes they are extended and separated from each other (open position). In some analyses, only CH1 binds to actin26 ; in others, both CH1 and CH2 bind.25,62 The EF hand domain is always in contact with the CH domains and not in contact with actin, so it presumably regulates the CH domains. The fact that α-actinin CH domains adopt both open and closed positions25 and utrophin also has 2 modes of binding63 suggests that the CH domains in spectrin might also do so and that this might be regulatory (convert weak binding to strong binding, for instance). Perhaps protein 4.1R and/or the EF domain act by converting the CH domains to a high-affinity conformation. Spectrin could occupy up to 2 actins in the actin filament, given the stoichiometry (6 spectrins per ∼14 actin monomers; see Table 1), so a variety of arrangements is possible. In any event, these are clearly key questions that need to be answered going forward.
Another obvious problem that is rarely mentioned is that spectrin is in a planar array, but the spectrin binding sites on the helical actin protofilament must be in a helical array. Spectrin does not hang down into the cytoplasm or stick up into the lipid bilayer, so if all the spectrin binding sites on actin are saturated, either the actin-binding end of spectrin must be flexible enough to curve around the actin filament, or spectrin must be able to bind to actin in more than 1 way.
What is the structure of spectrin on the membrane?
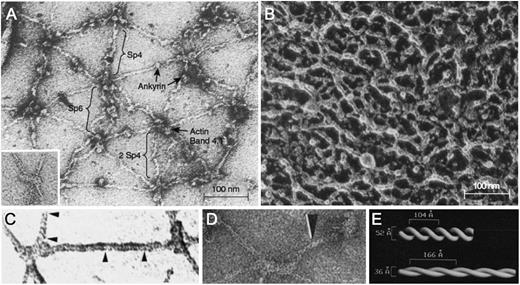
Electron micrographs of purified spectrin tetramers64 or spectrin filaments in fully stretched skeletons27 (Figure 5A 65 ) show a spaghetti-like molecule with an end-to-end length of up to 200 nm. This configuration is how spectrin is usually depicted in membrane models. However, the filaments of native, unstretched skeletons are much more compact (Figure 5B66 ), and simple calculations show that the average distance between actin filaments (ie, the length of a spectrin tetramer) is only 60 to 70 nm in vivo.6,67 This length corresponds roughly to that of presumed spectrin filaments in native skeletons.30,31,66 It is not known how spectrin folds in this resting state. The best evidence comes from visually enhanced electron micrographs of spectrin in partially expanded skeletons (Figure 5C-D), which suggest that α- and β-spectrin are coiled about each other in a 2-start helix with ∼10 turns per spectrin tetramer28 (Figure 5E). Some micrographs show relatively straight spectrin filaments28 ; others show some kinking.31 This difference may reflect whether the filaments are under tension. In this model, spectrin molecules extend and contract along the helical axis by varying the pitch and diameter of the double strand (Figure 5E).28 This springlike behavior is presumably critical to the elastic behavior of the membrane.
Negatively stained electron micrographs of red cell membrane skeletons and spectrin. (A) A membrane skeleton that was stretched during preparation. Note the pseudohexagonal organization of the skeleton and the location of various proteins. Inset: example of a 37-nm long F-actin protofilament. Sp4, spectrin tetramer; Sp6, spectrin hexamer; 2 Sp4, double spectrin tetramers. (B) Skeleton in situ in a red cell prepared by a minimally perturbing quick-freeze, deep-etch, rotary replication procedure. The dense network of filaments averages 29 ± 9 nm between intersections. (C-D) Spectrin from partially stretched skeletons. Some of the spectrin molecules show a helical periodic substructure, as noted by the region between the arrowheads in panel C or by the single arrowhead in panel D. (E) Right-handed double-helical models of spectrin periodicity obtained from the experiments in panel D by visual filtering of the periodic regions from multiple spectrin molecules. In this model, the springlike spectrins extend and contract by varying their pitch and diameter. Native spectrin tetramer has ∼10 turns, with a pitch of ∼7 nm (70 Å) and a diameter of ∼5.9 nm (59 Å). Panel A reprinted from Liu et al27 ; panel A inset from Byers and Branton65 ; panel B from Ursitti et al66 ; panel C from Ursitti and Wade30 ; and panels D and E from McGough and Josephs,28 all with permission.
Negatively stained electron micrographs of red cell membrane skeletons and spectrin. (A) A membrane skeleton that was stretched during preparation. Note the pseudohexagonal organization of the skeleton and the location of various proteins. Inset: example of a 37-nm long F-actin protofilament. Sp4, spectrin tetramer; Sp6, spectrin hexamer; 2 Sp4, double spectrin tetramers. (B) Skeleton in situ in a red cell prepared by a minimally perturbing quick-freeze, deep-etch, rotary replication procedure. The dense network of filaments averages 29 ± 9 nm between intersections. (C-D) Spectrin from partially stretched skeletons. Some of the spectrin molecules show a helical periodic substructure, as noted by the region between the arrowheads in panel C or by the single arrowhead in panel D. (E) Right-handed double-helical models of spectrin periodicity obtained from the experiments in panel D by visual filtering of the periodic regions from multiple spectrin molecules. In this model, the springlike spectrins extend and contract by varying their pitch and diameter. Native spectrin tetramer has ∼10 turns, with a pitch of ∼7 nm (70 Å) and a diameter of ∼5.9 nm (59 Å). Panel A reprinted from Liu et al27 ; panel A inset from Byers and Branton65 ; panel B from Ursitti et al66 ; panel C from Ursitti and Wade30 ; and panels D and E from McGough and Josephs,28 all with permission.
However, there is evidence for other models that propose that spectrin molecules adopt a random wormlike configuration,68 or that helical segments of the repeats melt and extend under tension,35 or simply unfold when stressed.36 The fact that previously occluded cysteines can be labeled when red cells are deformed by shear is evidence that some repeats unfold and that spectrin may even detach from ankyrin or actin when stretched.69 Because the structure of spectrin affects its mechanical properties, as well as the spatial relationships of all the other proteins that bind to it, defining its structure in vivo, at rest, and when deformed is a high priority.
Attachment of the membrane skeleton
Ankyrin
Erythrocyte ankyrin links spectrin to a complex of band 3 and other proteins in the lipid bilayer. The protein has 3 domains70 (Figure 6 70-73 ). The membrane domain is composed entirely of ankyrin repeats. It is a remarkably open, spiral structure, somewhat like a twisted sickle, that encircles and binds band 3 tetramers (Figure 6B 74 ). Whether band 3 spontaneously forms tetramers and then attracts ankyrin, stabilizing the tetramers, or whether ankyrin, which has 2 binding sites for band 3 (Figure 6A), independently binds 2 band 3 dimers (ie, a “dimer of dimers”), which then form a tetramer, is unresolved. The fact that a stable subpopulation of band 3 tetramers can be isolated from nonionic detergent extracts of ghosts75 argues for the former. But the structure of a band 3 tetramer binding to the full membrane domain has not been solved, and at least 1 recent paper argues that band 3 dimers must bind independently,76 so the question remains.
Ankyrin. (A) Schematic of erythroid ankyrin structure.70 The membrane domain contains 24 ankyrin repeats, grouped functionally into 4 subdomains of 6 repeats. There are 2 binding sites for band 3, one involving repeats 7 to 12 (domain D2), and one involving repeats 13 to 24 (D3/D4). The ankyrin binding loops on band 3 are predicted to interact with ankyrin repeats 19 and 20 (light blue) of D4.71 The interaction site within D2 is not known. The spectrin domain contains 3 subdomains, of which ZU5A (light blue) contains the binding site for spectrin.15,72 The C-terminal (regulatory) domain is thought to modulate the binding functions of the other 2 domains,70,73 and exists in numerous spliced variants of mostly unknown function. The function of the conserved death domain (DD) is also a mystery. (B) Hypothetical model74 of the interaction between the membrane domain of ankyrin (green) and a band 3 tetramer (red, blue, cyan, and purple subunits). Note how the ankyrin repeats form a large (9-nm diameter) twisted helical spiral. In this deduced model, the concave surface of the D3/D4 region interacts with the red subunit in 1 band 3 dimer. One subunit in the second band 3 dimer contacts the concave surface of the D2 region, which contains a second band 3 binding site. (C) Spectrin-ankyrin interaction. Note how ZU5A, the spectrin-binding subdomain within ankyrin, binds in the notch created by the sharp angle between spectrin repeats β14 and β15. Panel B reprinted from Michaely et al74 and panel C from Ipsaro and Mondragón,15 both with permission.
Ankyrin. (A) Schematic of erythroid ankyrin structure.70 The membrane domain contains 24 ankyrin repeats, grouped functionally into 4 subdomains of 6 repeats. There are 2 binding sites for band 3, one involving repeats 7 to 12 (domain D2), and one involving repeats 13 to 24 (D3/D4). The ankyrin binding loops on band 3 are predicted to interact with ankyrin repeats 19 and 20 (light blue) of D4.71 The interaction site within D2 is not known. The spectrin domain contains 3 subdomains, of which ZU5A (light blue) contains the binding site for spectrin.15,72 The C-terminal (regulatory) domain is thought to modulate the binding functions of the other 2 domains,70,73 and exists in numerous spliced variants of mostly unknown function. The function of the conserved death domain (DD) is also a mystery. (B) Hypothetical model74 of the interaction between the membrane domain of ankyrin (green) and a band 3 tetramer (red, blue, cyan, and purple subunits). Note how the ankyrin repeats form a large (9-nm diameter) twisted helical spiral. In this deduced model, the concave surface of the D3/D4 region interacts with the red subunit in 1 band 3 dimer. One subunit in the second band 3 dimer contacts the concave surface of the D2 region, which contains a second band 3 binding site. (C) Spectrin-ankyrin interaction. Note how ZU5A, the spectrin-binding subdomain within ankyrin, binds in the notch created by the sharp angle between spectrin repeats β14 and β15. Panel B reprinted from Michaely et al74 and panel C from Ipsaro and Mondragón,15 both with permission.
The ankyrin-spectrin binding site is well defined. The ZU5A subdomain of ankyrin binds to a special site created by the junction of the 14th and 15th repeats within β-spectrin (Figure 6C). Only 1 ankyrin binds per tetramer, perhaps for steric reasons. Ankyrin binding promotes spectrin tetramer and oligomer formation by ∼10-fold, and vice versa.16 Ankyrin attachment to band 3 also strengthens spectrin self-association.77 Presumably, both of these mechanisms help regulate the relatively tenuous self-association interaction.
Band 3
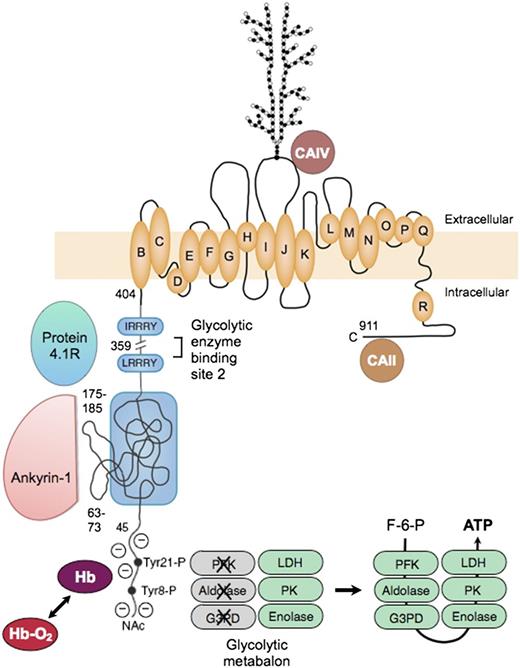
Erythrocyte band 3, or officially SLC4A1, is the major red cell membrane protein, with ∼1.2 million copies per cell. Functionally, it is 2 proteins78 : (1) an N-terminal cytoplasmic, peripheral membrane protein that is a key attachment site for the membrane skeleton, glycolytic enzymes, and deoxyhemoglobin; and (2) a C-terminal integral membrane protein that forms the red cell anion-exchange channel and aids carbon dioxide transport (Figure 7 78-86 ).
Organizational model of human erythrocyte band 3 (anion-exchange protein). The protein contains 2 structurally and functionally distinct domains: a cytoplasmic binding domain (amino acids 1-359) and a transmembrane domain (amino acids 360-911) that forms the anion-exchange channel.78 In the cytoplasmic domain, the glycolytic enzymes PFK, aldolase, and G3PD bind to amino acids 1 to 23 at the N-terminus of band 3 and contact amino acids 356 to 384,79 which are nearby in the folded protein. The enzymes are inactive when bound,80 but are displaced and activated by deoxyhemoglobin (Hb), which also binds to the N-terminus,81 or by phosphorylation of 2 tyrosines (Tyr21-P and Tyr8-P) within the binding sites.80 Enolase, PK and LDH also localize to the membrane and are displaced by deoxyhemoglobin, but do not bind to band 3.80 This fact suggests that many of the enzymes in the glycolytic pathway form a functional complex (or “metabolon”) that efficiently generates adenosine triphosphate (ATP), particularly under hypoxic conditions. Ankyrin also interacts with the N-terminus,82 but the main ankyrin sites are amino acids 63-73 and 175-185, which loop out from the surface.71 Protein 4.1 binds to 2 sites with the (I/L)RRRY motif, near the end of the domain.83 Ankyrin and protein 4.1R inhibit each other’s binding. The binding sites for protein 4.2 and adducin have not been identified. In the membrane domain, the best current model84 for the anion-exchange channel is based on the structure of the ClC bacterial chloride channel. The intramembrane and transmembrane helices are lettered as they are in the ClC channel. A complex carbohydrate structure is attached to Asn 642. Carbonic anhydrase (CA)IV binds to the extracellular loop of band 3 between helices I and J,85 and CAII may bind to the C-terminal segment86 ; both are perfectly positioned to create bicarbonate from carbon dioxide and to shuttle the ions to or from the plasma through band 3. F-6-P, fructose 6-phosphate; Hb-O2, oxyhemoglobin; NAc, acetylated aminoterminus.
Organizational model of human erythrocyte band 3 (anion-exchange protein). The protein contains 2 structurally and functionally distinct domains: a cytoplasmic binding domain (amino acids 1-359) and a transmembrane domain (amino acids 360-911) that forms the anion-exchange channel.78 In the cytoplasmic domain, the glycolytic enzymes PFK, aldolase, and G3PD bind to amino acids 1 to 23 at the N-terminus of band 3 and contact amino acids 356 to 384,79 which are nearby in the folded protein. The enzymes are inactive when bound,80 but are displaced and activated by deoxyhemoglobin (Hb), which also binds to the N-terminus,81 or by phosphorylation of 2 tyrosines (Tyr21-P and Tyr8-P) within the binding sites.80 Enolase, PK and LDH also localize to the membrane and are displaced by deoxyhemoglobin, but do not bind to band 3.80 This fact suggests that many of the enzymes in the glycolytic pathway form a functional complex (or “metabolon”) that efficiently generates adenosine triphosphate (ATP), particularly under hypoxic conditions. Ankyrin also interacts with the N-terminus,82 but the main ankyrin sites are amino acids 63-73 and 175-185, which loop out from the surface.71 Protein 4.1 binds to 2 sites with the (I/L)RRRY motif, near the end of the domain.83 Ankyrin and protein 4.1R inhibit each other’s binding. The binding sites for protein 4.2 and adducin have not been identified. In the membrane domain, the best current model84 for the anion-exchange channel is based on the structure of the ClC bacterial chloride channel. The intramembrane and transmembrane helices are lettered as they are in the ClC channel. A complex carbohydrate structure is attached to Asn 642. Carbonic anhydrase (CA)IV binds to the extracellular loop of band 3 between helices I and J,85 and CAII may bind to the C-terminal segment86 ; both are perfectly positioned to create bicarbonate from carbon dioxide and to shuttle the ions to or from the plasma through band 3. F-6-P, fructose 6-phosphate; Hb-O2, oxyhemoglobin; NAc, acetylated aminoterminus.
The glycolytic enzymes form a metabolic complex (metabolon) extending from phosphofructokinase through lactic dehydrogenase. Three of the enzymes bind to the N-terminus of band 3: phosphofructokinase, aldolase, and glyceraldehyde-3-phosphate dehydrogenase. The others bind indirectly as part of the complex.80 The enzymes are inactive when bound80 but are displaced and activated either by deoxyhemoglobin, which also binds to the N-terminus,81 or by phosphorylation of 2 tyrosines within the binding sites80 (Figure 7). The enzymes80 and their adenosine triphosphate product87 are located in discrete clumps along the membrane, supporting the idea of a physical metabolon. Conditions that displace the enzymes from the membrane activate glycolytic fluxes in intact red cells by 45%, which supports the idea of a functional metabalon.88
As noted earlier, ankyrin binds to band 3 tetramers. The acidic N-terminus and 2 loops on the surface of band 3 are involved in the binding69 (Figure 7). As with the glycolytic metabolon, ankyrin is displaced from band 3 by deoxyhemoglobin.82 The affinity of deoxyhemoglobin is weak, but the concentration of hemoglobin is so high in red cells that approximately half of the band 3 molecules are bound to deoxyhemoglobin when red cells are deoxygenated.82 Loosened ankyrin constraints could improve blood flow in hypoxic areas, but with the risk that red cells subject to prolonged deoxygenation, such as those trapped in the spleen, might suffer membrane vesiculation. (Could this be the long-sought mechanism for splenic conditioning89 in hereditary spherocytosis?).
The ankyrin complex
Ankyrin and band 3 are part of the multiprotein ankyrin complex (Figure 2). From the amounts of each protein in the red cell (Table 1), the complex is estimated to contain 1 ankyrin, 1 band 3 tetramer, 2 glycophorin A or B dimers or heterodimers, 2 protein 4.2 molecules, and 1 Rh complex (a trimer of RhAG combined with RhD and RhCE). CD47, the Landsteiner-Wiener blood group antigen, and the proteins in the glycolytic metabolon are also components, but are present in less than stoichiometric amounts (Table 1) and so must contribute only to a subset of complexes. One wonders how many such subsets there are. Do they have constant composition or are they in a dynamic equilibrium? And are the rare complexes that contain CD47 and Landsteiner-Wiener blood group antigen localized to specific membrane subdomains?
As detailed in Table 1 and Figure 2, there are multiple interactions among proteins in the ankyrin complex. Some of these are critical; for example, red cells lacking band 3 also lack protein 4.2 and glycophorin A,90 and red cells lacking protein 4.2 nearly lack CD47.91 A consequence of this promiscuity is that missense mutations that interfere with a single interaction within the ankyrin complex have little effect. Hereditary spherocytosis is caused by defects in spectrin, ankyrin, band 3, or protein 4.2 that impair ankyrin complex formation, and almost all the known mutations diminish the concentration of these proteins rather than block their binding functions.
The actin junctional complex
The actin protofilament and its associated proteins are also attached to the membrane. For many years, this attachment was believed to occur exclusively via a ternary complex of protein 4.1R, p55, and glycophorin C or D. Each of these proteins interacts with the other 2, and the interaction sites are well mapped.48,92,93 However, if this was an important skeleton attachment and the sole attachment at the actin-binding end of spectrin, it was never clear why patients with complete absence of glycophorin C and D had, at most, very mild hereditary elliptocytosis.
During the past few years, it has become clear that the actin junctional complex is more complex and, like the ankyrin complex, is centered on band 3 (Figure 2). First, protein 4.1R and ankyrin compete for binding to band 3,94 which suggests that they bind to separate band 3 populations. The interaction with protein 4.1R is known to be important because zebrafish lacking band 3 can be rescued by expression of mouse band 3 but only if the mouse band 3 contains the protein 4.1R binding sites.95 Second, protein 4.2, which requires band 3 for incorporation into the membrane, binds to the EF hand domain at the actin-binding end of spectrin. Third, adducin binds to band 3,57 which definitely localizes a subpopulation of band 3 to the neighborhood of actin (Figure 2). The latter interaction is important because the membrane fragments when the connection is severed.57
The stoichiometry of the actin junctional complex is uncertain. On average, 6 spectrins and 6 proteins 4.1R bind per actin (Figure 8 96 ), and there is enough band 3, glycophorin A, glycophorin C/D, and probably enough glucose transporter 1 and stomatin to supply 6 complexes per actin filament (Table 1). However, there is only 1 copy of adducin per actin filament and only 3 copies of dematin, if it is a monomer as recent studies indicate,59 or one copy if it is a trimer. Similarly, whereas protein 4.1R, glycophorin C/D, and protein p55 are usually pictured as a 3-part complex, there is only enough p55 for 1 copy per actin filament if it is a dimer, as some data suggest.9 7 And there is only ∼1 copy of protein 4.2 available if 240 000 copies (1 per band 3 dimer) are tied up in the ankyrin complex. The actin junctional complex also contains enzymes in the glycolytic metabolon,79 as well as the blood group proteins Kx/Kell and DARC/Duffy,98 although the amounts of these proteins are insufficient for even 1 copy per complex.
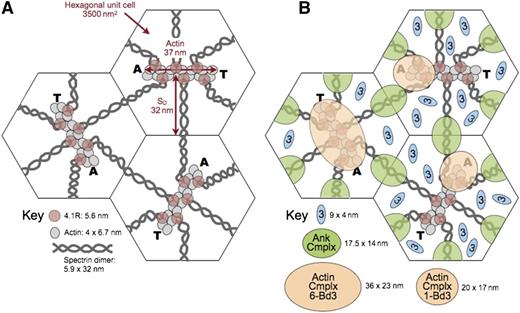
Model of the unit cell of the membrane skeleton. (A) The membrane skeleton is a quasihexagonal lattice centered around ∼40 000 F-actin protofilaments. Each 37-nm, double-helical protofilament is capped by adducin (A) and tropomodulin (T) and contains ∼14 G-actin subunits. The red cell surface area is ∼140 μm2, so each hexagonal unit cell is 3500 nm2 and the average length of a spectrin dimer is ∼32 nm, which is one-third its length when fully stretched. Available data suggest that the spectrin chains are coiled about each other and expand and contract by changing their pitch and diameter28 (Figure 5E). They are shown here as straight, although they are sometimes kinked.31 Spectrin oligomers are not shown in this model but exist in vivo. In addition, the skeleton is not as regularly arrayed in vivo as in this averaged model. (B) A recent model of the ankyrin complex (Ank Cmplx; green ovals) assembled from known structures of the proteins by Burton and Bruce7 has a cross-sectional area of 17.5 nm × 14 nm, assuming the proteins are closely packed. The cross-sectional area of the actin junctional complex is estimated to be ∼20 nm × 17 nm if it contains only a single band 3 dimer7 (Actin Cmplx 1-Bd3; small tan ovals). Such a complex would presumably lie toward the end of the actin protofilament where adducin resides. This position is shown in the 2 unit cells on the right. Alternatively, if the actin junctional complex contains 6 band 3 dimers (Actin Cmplx 6-Bd3) interacting with all 6 proteins 4.1R, it would probably be more centered and approximate the area of the large tan oval in the unit cell on the left. Recent data favor the larger arrangement,96 but in either case, it is likely that the ankyrin and actin junctional complexes would be close to each other and probably often touch, particularly during red cell deformation. Band 3 dimers that are not associated with either the ankyrin or actin junctional complexes (small blue ovals) diffuse in the lipid bilayer within the spectrin corrals. There are 3 to 8 of these per unit cell, depending on the number of band 3 dimers in the actin junctional complex. Reprinted from Lux6 with permission.
Model of the unit cell of the membrane skeleton. (A) The membrane skeleton is a quasihexagonal lattice centered around ∼40 000 F-actin protofilaments. Each 37-nm, double-helical protofilament is capped by adducin (A) and tropomodulin (T) and contains ∼14 G-actin subunits. The red cell surface area is ∼140 μm2, so each hexagonal unit cell is 3500 nm2 and the average length of a spectrin dimer is ∼32 nm, which is one-third its length when fully stretched. Available data suggest that the spectrin chains are coiled about each other and expand and contract by changing their pitch and diameter28 (Figure 5E). They are shown here as straight, although they are sometimes kinked.31 Spectrin oligomers are not shown in this model but exist in vivo. In addition, the skeleton is not as regularly arrayed in vivo as in this averaged model. (B) A recent model of the ankyrin complex (Ank Cmplx; green ovals) assembled from known structures of the proteins by Burton and Bruce7 has a cross-sectional area of 17.5 nm × 14 nm, assuming the proteins are closely packed. The cross-sectional area of the actin junctional complex is estimated to be ∼20 nm × 17 nm if it contains only a single band 3 dimer7 (Actin Cmplx 1-Bd3; small tan ovals). Such a complex would presumably lie toward the end of the actin protofilament where adducin resides. This position is shown in the 2 unit cells on the right. Alternatively, if the actin junctional complex contains 6 band 3 dimers (Actin Cmplx 6-Bd3) interacting with all 6 proteins 4.1R, it would probably be more centered and approximate the area of the large tan oval in the unit cell on the left. Recent data favor the larger arrangement,96 but in either case, it is likely that the ankyrin and actin junctional complexes would be close to each other and probably often touch, particularly during red cell deformation. Band 3 dimers that are not associated with either the ankyrin or actin junctional complexes (small blue ovals) diffuse in the lipid bilayer within the spectrin corrals. There are 3 to 8 of these per unit cell, depending on the number of band 3 dimers in the actin junctional complex. Reprinted from Lux6 with permission.
Therefore, it is uncertain at the moment whether the actin junctional complex contains only a single band 3 dimer, interacting with just 1 or 2 of the 6 spectrin/protein 4.1R dyads, or whether it is a larger complex containing 3 to 6 band 3 dimers that interact with all the spectrins and proteins 4.1R. Both possibilities are depicted in Figure 8. Although the data are not conclusive, recent evidence favors a larger complex.96 If so, it is not clear whether all or just a subset of band 3 molecules bind to adducin.
In either case, as shown in Figure 8, an interesting and underappreciated fact is that the ankyrin and actin junctional complexes are quite close to each other in situ on the membrane and must often collide, especially during red cell deformation but perhaps even when the cell is at rest. This knowledge raises the interesting possibility that proteins like band 3, protein 4.1R, protein 4.2, and adducin, which have binding partners in both complexes, may sometimes switch allegiances. Because some proteins compete for the same binding sites (eg, as noted, protein 4.1R displaces ankyrin from band 3),94 this process could be a regulatory one.
Distribution of band 3
Of the ∼1.2 million band 3 monomers per red cell, 40% are tetramers bound to ankyrin.96 Depending on whether the actin junctional complex contains an average of 1 or 6 band 3 dimers, an additional 7% to 40% of the band 3 dimers are located there. Recent experiments comparing normal mouse erythrocytes to α-adducin-deficient erythrocytes suggest that roughly 33% of the band 3 molecules are immobilized by adducin, which favors a larger complex.96 The remaining band 3 dimers (25%-30%) are believed to be diffusing freely in the lipid bilayer, constrained for the most part by the boundaries of the spectrin corrals, which average <100 nm.96 These percentages are remarkably similar to those of freely diffusing band 3 dimers (25%-29%) measured in normal red cells using different methods.96,99,100
It is important to remember that some of the minor proteins found in band 3 complexes can exist only in a subset of the complexes (probably multiple different subsets) and that the lipid bilayer is heterogeneous, with areas like the lipid rafts where certain proteins concentrate. Therefore, the band 3 complexes must be more heterogeneous than these 3 states of band 3 imply. Analyses of band 3 mobilities support this conclusion.95
The future
A great deal has been learned about the structure of the red cell membrane and the membrane skeleton in the 36 years since the first model was published (Figure 1). I sometimes hear people say that the red cell membrane is no longer an active area of hematology research, but I disagree. We have the broad outlines of how the membrane is organized (Figure 2) but know few of the details, and what we do know is about a static structure. Many questions remain (Table 2). We do not know how the skeleton is constructed during erythropoiesis. We know very little about the dynamics of the skeleton: how labile individual bonds are, how the skeleton responds to capillary deformation, how the skeleton is disassembled and reassembled during invasion by malaria or other parasites, and so forth. And as discussed earlier, we do not know how spectrin is folded in the intact skeleton or how the proteins that form the actin junctional complex are organized. Or how spectrin binds to actin. We do not know whether band 3–binding proteins like protein 4.1R, protein 4.2, and adducin switch from one band 3 complex to another, given the apparent proximity of the complexes at each end of spectrin (Figure 8). We do not understand why the same repeats in different spectrins are so similar if they just serve as spacers. Do they have binding or mechanical functions that we do not yet know about? We do not know what roles posttranslational changes like phosphorylation, fatty acylation, methylation, glycation, hydroxylation, and ubiquitination play, or what roles calcium ion–binding and adenosine triphosphate–binding play. And we do not understand the interactions between the membrane skeleton and the overlying lipids, which will likely have regulatory as well as structural functions. This is only a partial list of questions; some others are itemized in Table 2. But I hope my point is clear. Because the red cell membrane skeleton is the paradigm for studies of spectrin-based membrane skeletons in all cells, and because we now know that spectrin and other membrane skeletal proteins invest internal organelles, some transport vesicles, and plasma membranes, and even play a role in nuclear organization, it is more important than ever to understand the structure and function of the red cell membrane skeleton.
Some unanswered questions
| General | What are the amounts and stoichiometries of the membrane proteins (only a few are accurately measured)? |
| Spectrin | What is the structure of spectrin in vivo? How does it change when the membrane is deformed? |
| How does spectrin bind to the actin protofilament? What is the stoichiometry? Where do the CH1 and CH2 domains and protein 4.1R bind on actin? Is there >1 binding conformation? If so, how do these differ functionally and how are they regulated? | |
| Why is the EF hand domain essential for normal spectrin-actin binding? How does it work? | |
| How do spectrin molecules, which lie in a plane, accommodate different orientations of their binding sites on the helical actin protofilament? | |
| Higher order spectrin oligomers (hexamers, octamers, etc) also serve as molecular junctions in the skeleton. Are there significant numbers of these and do they have a unique functional role? | |
| If the available spectrin repeat structures are correct and there is a continuous α-helix across the interrepeat junction, how is spectrin flexibility achieved? | |
| Why are the same repeats in different spectrins so similar? Do they have binding or mechanical functions that we do not yet know about? | |
| What binds to the spectrin src homology 3 (SH3) domain? What are the consequences? | |
| There is evidence that some of the spectrins in mature red cells contain the muscle isoform of β1-spectrin, which has a lipid-interacting pleckstrin homology domain. Is this true, and if so, what is its function? | |
| Actin | How are actin protofilaments formed? How do they achieve their uniform size? Do formins play a role? |
| Are the actin protofilaments stable or do they turn over, as the presence of a critical concentration of G-actin in the red cell cytoplasm suggests? If so, how is this regulated? | |
| How are the various proteins arranged on the actin protofilament? Is the arrangement the same in all the protofilaments? If there are open binding sites on actin, do molecules like spectrin move from site to site with membrane deformation? | |
| Do red cell myosin and caldesmon have a function in the membrane of mature red cells? | |
| Ankyrin | Does ankyrin bind a band 3 tetramer or 2 dimers? If a tetramer, is it preformed before binding or do 2 dimers bind and convert to a tetramer on ankyrin. What is the structure of the band 3 tetramer-ankyrin complex? |
| What is the structure of the ankyrin molecule? How do the various domains relate to each other spatially? | |
| Where do protein 4.2 and RhAG bind? | |
| What is the function of the death domain in ankyrin? | |
| What are the functions of the many ankyrin spliceoforms? | |
| Band 3 | What is the overall structure of band 3? |
| Where do protein 4.2 and adducin bind on band 3? | |
| Because the numbers of integral membrane proteins vary by orders of magnitude (Table 1), the band 3 multiprotein complexes must vary in composition. How many such complexes are there? Do they have specific, stable compositions or are they in equilibrium and continually changing their composition? Are the substoichiometric proteins (eg, CD44, CD47, LW, Kx/Kell, DARC) uniformly distributed or localized to lipid rafts or other membrane lipid or protein subdomains? | |
| How are the proteins in the band 3 multiprotein complexes arranged relative to each other? | |
| Do protein interactions within the band 3 multiprotein complexes allosterically affect the interactions or functions of other proteins in the complexes? | |
| Does protein 4.1R bind to band 3 in vivo? If so, does band 3 bind to each of the spectrin/actin/protein 4.1R complexes or only to a subset, such as the subset that interacts with adducin (Figure 8)? | |
| Are any band 3 molecules “unbound” (ie, not attached to the membrane skeleton directly or indirectly). If so, how many? Are they also part of multiprotein complexes? Do they have a unique function? | |
| Glycolytic metabolon | The enzymes that compose the proposed glycolytic metabolon vary in concentration by orders of magnitude. Are the estimates correct? If so, do some complexes lack rare components (but then, how would they function?) or are they gigantic so as to include at least 1 copy of each enzyme? Are other enzymes in the glycolytic pathway part of the complex? |
| How is the glycolytic metabolon regulated in vivo? What is the purpose of activating glycolysis by deoxyhemoglobin? Do products of glycolysis such as adenosine triphosphate (ATP) cause vasodilation directly or indirectly? Does nitric oxide play a role? Is the rate of dissociation and activation of the glycolytic metabolon rapid enough to be relevant during normal circulatory transits? | |
| Protein 4.1R | What is the structure of intact protein 4.1R and the spectrin/actin/protein 4.1R complex? |
| Calmodulin binds to and regulates protein 4.1R. The EF hand domain is a calmodulin-like structure that resides near protein 4.1R in the actin junctional complex. In addition, the EF domain binds calmodulin. Does protein 4.1R bind to the EF domain or do they bind each other through calmodulin? If so, is this a regulatory interaction? | |
| Does phosphatidyl-4,5-bisphosphate regulate protein 4.1R binding to actin or other proteins in vivo? | |
| Because there is insufficient protein p55/palmitoylated membrane protein to make a 1:1:1 molar complex with protein 4.1R and glycophorin C/D, how is the complex constructed? | |
| Protein 4.2 | What is the function of protein 4.2 when attached to the spectrin EF hand domain? Does it actually occupy the domain in vivo? If so, does it bind to protein 4.1R? |
| What is the function of the ATP-binding site on protein 4.2? | |
| Adducin | Is adducin a dimer or a tetramer? |
| What is the structure of adducin? How does the C-terminal tail relate to the head end? | |
| Band 3, spectrin, and actin all bind to the C-terminal tail. What is the structure of this complex? How are the multiple interactions regulated? Can they occur all at once on the same molecule or do some interactions interfere with others? When adducin binds spectrin and actin, does it also interact with nearby proteins 4.1R or 4.2? Does adducin bind band 3 dimers or tetramers, and does it bind 1 or 2 molecules of each? | |
| Dematin | How does dematin contribute to spectrin binding in vivo? |
| Is the ability of dematin to bundle actin filaments important in the mature red cell? | |
| Where does dematin bind relative to spectrin and adducin on the actin protofilament? Does the number of dematins per protofilament vary? | |
| Tropomyosin | Do the red cell isoforms of tropomyosin determine the unique structure of the short actin protofilaments? Do specific actin-nucleating formins play a role in this process? |
| Posttranslational modifications | What do posttranslational modifications like phosphorylation, palmitoylation, myristoylation, hydroxylation, methylation, glycation, and ubiquitination do? Are the effects important in vivo? |
| How are the multiple effects of calcium ion and calmodulin integrated in vivo? | |
| Dynamics | Do the actin junctional complex and ankyrin complex contact each other in vivo? Do proteins like band 3, proteins 4.1R and 4.2, and adducin, which have potential binding partners in both complexes, sometimes switch allegiances? If so, is this a regulatory process? |
| How labile are the individual bonds in the membrane skeleton? Which bonds, other than the spectrin dimer-tetramer interaction, break when the skeleton is deformed? Are the rates of dissociation and reassociation such that the bonds dissociate during red cell deformation in the circulation? | |
| How is the skeleton disassembled and reassembled during invasion by malaria and other parasites? | |
| Lipids | What are the interactions between the membrane skeleton and the overlying lipids? Do these interactions have regulatory as well as structural functions? Does the membrane skeleton vary in unique regions of the lipid bilayer such as lipid rafts? |
| General | What are the amounts and stoichiometries of the membrane proteins (only a few are accurately measured)? |
| Spectrin | What is the structure of spectrin in vivo? How does it change when the membrane is deformed? |
| How does spectrin bind to the actin protofilament? What is the stoichiometry? Where do the CH1 and CH2 domains and protein 4.1R bind on actin? Is there >1 binding conformation? If so, how do these differ functionally and how are they regulated? | |
| Why is the EF hand domain essential for normal spectrin-actin binding? How does it work? | |
| How do spectrin molecules, which lie in a plane, accommodate different orientations of their binding sites on the helical actin protofilament? | |
| Higher order spectrin oligomers (hexamers, octamers, etc) also serve as molecular junctions in the skeleton. Are there significant numbers of these and do they have a unique functional role? | |
| If the available spectrin repeat structures are correct and there is a continuous α-helix across the interrepeat junction, how is spectrin flexibility achieved? | |
| Why are the same repeats in different spectrins so similar? Do they have binding or mechanical functions that we do not yet know about? | |
| What binds to the spectrin src homology 3 (SH3) domain? What are the consequences? | |
| There is evidence that some of the spectrins in mature red cells contain the muscle isoform of β1-spectrin, which has a lipid-interacting pleckstrin homology domain. Is this true, and if so, what is its function? | |
| Actin | How are actin protofilaments formed? How do they achieve their uniform size? Do formins play a role? |
| Are the actin protofilaments stable or do they turn over, as the presence of a critical concentration of G-actin in the red cell cytoplasm suggests? If so, how is this regulated? | |
| How are the various proteins arranged on the actin protofilament? Is the arrangement the same in all the protofilaments? If there are open binding sites on actin, do molecules like spectrin move from site to site with membrane deformation? | |
| Do red cell myosin and caldesmon have a function in the membrane of mature red cells? | |
| Ankyrin | Does ankyrin bind a band 3 tetramer or 2 dimers? If a tetramer, is it preformed before binding or do 2 dimers bind and convert to a tetramer on ankyrin. What is the structure of the band 3 tetramer-ankyrin complex? |
| What is the structure of the ankyrin molecule? How do the various domains relate to each other spatially? | |
| Where do protein 4.2 and RhAG bind? | |
| What is the function of the death domain in ankyrin? | |
| What are the functions of the many ankyrin spliceoforms? | |
| Band 3 | What is the overall structure of band 3? |
| Where do protein 4.2 and adducin bind on band 3? | |
| Because the numbers of integral membrane proteins vary by orders of magnitude (Table 1), the band 3 multiprotein complexes must vary in composition. How many such complexes are there? Do they have specific, stable compositions or are they in equilibrium and continually changing their composition? Are the substoichiometric proteins (eg, CD44, CD47, LW, Kx/Kell, DARC) uniformly distributed or localized to lipid rafts or other membrane lipid or protein subdomains? | |
| How are the proteins in the band 3 multiprotein complexes arranged relative to each other? | |
| Do protein interactions within the band 3 multiprotein complexes allosterically affect the interactions or functions of other proteins in the complexes? | |
| Does protein 4.1R bind to band 3 in vivo? If so, does band 3 bind to each of the spectrin/actin/protein 4.1R complexes or only to a subset, such as the subset that interacts with adducin (Figure 8)? | |
| Are any band 3 molecules “unbound” (ie, not attached to the membrane skeleton directly or indirectly). If so, how many? Are they also part of multiprotein complexes? Do they have a unique function? | |
| Glycolytic metabolon | The enzymes that compose the proposed glycolytic metabolon vary in concentration by orders of magnitude. Are the estimates correct? If so, do some complexes lack rare components (but then, how would they function?) or are they gigantic so as to include at least 1 copy of each enzyme? Are other enzymes in the glycolytic pathway part of the complex? |
| How is the glycolytic metabolon regulated in vivo? What is the purpose of activating glycolysis by deoxyhemoglobin? Do products of glycolysis such as adenosine triphosphate (ATP) cause vasodilation directly or indirectly? Does nitric oxide play a role? Is the rate of dissociation and activation of the glycolytic metabolon rapid enough to be relevant during normal circulatory transits? | |
| Protein 4.1R | What is the structure of intact protein 4.1R and the spectrin/actin/protein 4.1R complex? |
| Calmodulin binds to and regulates protein 4.1R. The EF hand domain is a calmodulin-like structure that resides near protein 4.1R in the actin junctional complex. In addition, the EF domain binds calmodulin. Does protein 4.1R bind to the EF domain or do they bind each other through calmodulin? If so, is this a regulatory interaction? | |
| Does phosphatidyl-4,5-bisphosphate regulate protein 4.1R binding to actin or other proteins in vivo? | |
| Because there is insufficient protein p55/palmitoylated membrane protein to make a 1:1:1 molar complex with protein 4.1R and glycophorin C/D, how is the complex constructed? | |
| Protein 4.2 | What is the function of protein 4.2 when attached to the spectrin EF hand domain? Does it actually occupy the domain in vivo? If so, does it bind to protein 4.1R? |
| What is the function of the ATP-binding site on protein 4.2? | |
| Adducin | Is adducin a dimer or a tetramer? |
| What is the structure of adducin? How does the C-terminal tail relate to the head end? | |
| Band 3, spectrin, and actin all bind to the C-terminal tail. What is the structure of this complex? How are the multiple interactions regulated? Can they occur all at once on the same molecule or do some interactions interfere with others? When adducin binds spectrin and actin, does it also interact with nearby proteins 4.1R or 4.2? Does adducin bind band 3 dimers or tetramers, and does it bind 1 or 2 molecules of each? | |
| Dematin | How does dematin contribute to spectrin binding in vivo? |
| Is the ability of dematin to bundle actin filaments important in the mature red cell? | |
| Where does dematin bind relative to spectrin and adducin on the actin protofilament? Does the number of dematins per protofilament vary? | |
| Tropomyosin | Do the red cell isoforms of tropomyosin determine the unique structure of the short actin protofilaments? Do specific actin-nucleating formins play a role in this process? |
| Posttranslational modifications | What do posttranslational modifications like phosphorylation, palmitoylation, myristoylation, hydroxylation, methylation, glycation, and ubiquitination do? Are the effects important in vivo? |
| How are the multiple effects of calcium ion and calmodulin integrated in vivo? | |
| Dynamics | Do the actin junctional complex and ankyrin complex contact each other in vivo? Do proteins like band 3, proteins 4.1R and 4.2, and adducin, which have potential binding partners in both complexes, sometimes switch allegiances? If so, is this a regulatory process? |
| How labile are the individual bonds in the membrane skeleton? Which bonds, other than the spectrin dimer-tetramer interaction, break when the skeleton is deformed? Are the rates of dissociation and reassociation such that the bonds dissociate during red cell deformation in the circulation? | |
| How is the skeleton disassembled and reassembled during invasion by malaria and other parasites? | |
| Lipids | What are the interactions between the membrane skeleton and the overlying lipids? Do these interactions have regulatory as well as structural functions? Does the membrane skeleton vary in unique regions of the lipid bilayer such as lipid rafts? |
Acknowledgments
The author regrets that important papers from many talented individuals who have made significant contributions to red cell research could not be discussed or cited due to space limitations. The author gratefully acknowledges the camaraderie and friendship of a large number of colleagues in the red cell membrane field who, over many years, have enriched his career and aided his research.
Authorship
Contribution: S.E.L. wrote the review and designed the figures and tables.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Samuel E. Lux, Boston Children’s Hospital, 300 Longwood Ave, Hunnewell 260.1, Boston, MA 02115; e-mail: lux@enders.tch.harvard.edu.