In this issue of Blood, Nadeu et al delve deep into the genomes of patients diagnosed with chronic lymphocytic leukemia (CLL) and report on the clinical implications of tumor heterogeneity in the era of next-generation sequencing (NGS).1 While clonal evolution is common in hematologic tumors, it remains challenging to detect; however, this difficulty can be circumvented by performing ultradeep sequencing, which substantially improves the discovery of variants across a range of variant allelic frequencies (VAFs). This strategy was adopted by the Spanish group, which enabled them to not only detect subclonal mutations but also demonstrate that the presence of subclonal mutations within certain genes (ie, TP53 and NOTCH1) is clinically relevant.1
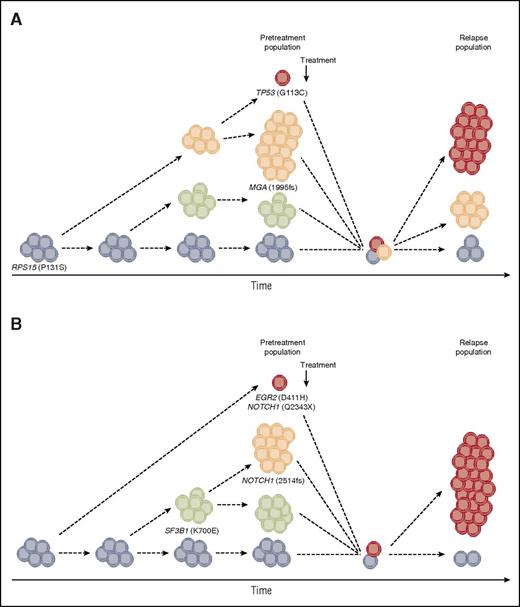
Clonal evolution and intraclonal dynamics in 2 CLL patients relapsing after fludarabine, cyclophosphamide, and rituximab therapy. Examples based on whole-exome sequencing data from our recent study (Ljungström et al).5 (A) In addition to the founding clone that harbors a RPS15 mutation, 2 subclones were observed before treatment initiation, one of which carried a frameshift deletion within MGA. At relapse, the MGA-deleted subclone disappeared, while a new subclone containing a TP53 mutation emerged. (B) A marked shift in subclonal populations was observed; while 2 subclones harboring the classical 2-bp NOTCH1 deletion and the recurrent p.K700E mutation in SF3B1 were eradicated after treatment, a new subclone emerged harboring a stop-gain NOTCH1 mutation and an EGR2 mutation.
Clonal evolution and intraclonal dynamics in 2 CLL patients relapsing after fludarabine, cyclophosphamide, and rituximab therapy. Examples based on whole-exome sequencing data from our recent study (Ljungström et al).5 (A) In addition to the founding clone that harbors a RPS15 mutation, 2 subclones were observed before treatment initiation, one of which carried a frameshift deletion within MGA. At relapse, the MGA-deleted subclone disappeared, while a new subclone containing a TP53 mutation emerged. (B) A marked shift in subclonal populations was observed; while 2 subclones harboring the classical 2-bp NOTCH1 deletion and the recurrent p.K700E mutation in SF3B1 were eradicated after treatment, a new subclone emerged harboring a stop-gain NOTCH1 mutation and an EGR2 mutation.
Enabled by technological advances, the full compendium of common, recurrent somatic mutations in the coding genome of CLL is nearing completion and has revealed that this malignancy is not characterized by a single unifying pathological mutation. Instead, a relatively restricted number of genes are mutated at a high frequency in CLL (TP53, ATM, NOTCH1, and SF3B1), with most recurrently affected genes located in the so-called long tail of the mutation distribution, occurring at a low frequency (<5%).2,3 We have also recently begun to understand the clinical impact of these mutations, with a number of genes harboring alterations linked to poor clinical outcome and enriched in patients with a clinically aggressive or chemorefractory disease (eg, NOTCH1, ATM, SF3B1, NFKBIE, BIRC3, and RPS15).4,5 In a recent multicenter study conducted by the European Research Initiative on CLL, the impact of TP53, NOTCH1, SF3B1, BIRC3, and MYD88 mutations were investigated in 3490 patients, and the study revealed that TP53 and SF3B1 mutations, but not NOTCH1 mutations, remained as independent markers of short time to first treatment (TTFT) in multivariate analysis.6 Studies have also suggested a predictive role for additional genes harboring mutations, but these have to be studied further before firm conclusions can be reached.
The development of NGS has changed the comprehensiveness of human genetic analysis and provided us with insights into subclonal complexity and the dynamic changes that occur over time and in relation to therapy (see figure).7 Recent studies have revealed that minor TP53 subclones (ie, subclones with a VAF not detected by Sanger sequencing) are as equally important to detect as clonal TP53 mutations (ie, detected by Sanger sequencing), because CLL patients harboring minor subclones at diagnosis exhibited a poor outcome similar to patients carrying clonal TP53 mutations.8,9 Nevertheless, the impact of recurrent mutations in other genes deemed to be potential “drivers” has not yet been investigated.
To clarify this matter, Nadeu et al performed targeted ultradeep NGS in a large, well-characterized cohort of 406 untreated CLL patients and investigated TP53, ATM, BIRC3, SF3B1, and NOTCH1.1 The authors initially made a series of important validation experiments to ensure high sequence depth/coverage and reproducibility and to determine the minimal VAF cutoff at which they could detect a mutation with their NGS strategy (range, 0.3% to 2%, depending on the specific gene analyzed). Using this approach, they identified a substantial proportion of subclonal mutations within TP53 (4.2%), SF3B1 (5.2%), and NOTCH1 (7.6%), while fewer subclonal ATM (1.3%) and BIRC3 (2.2%) mutations were observed. Confirming the known clinical impact of clonal mutations within TP53, ATM, NOTCH1, SF3B1, and BIRC3, they proceeded to demonstrate that CLL patients possessing subclonal NOTCH1 mutations had a significantly shorter TTFT, while subclonal TP53 mutations were associated with a shorter overall survival (OS). However, subclonal SF3B1 mutations did not appear to influence disease outcome (too few subclonal ATM and BIRC3 mutations were detected to allow for a meaningful analysis). Genomic analyses also permits inference about the temporality of mutational events, and by monitoring a subset of patients longitudinally, Nadeu et al were able to gain insight into the complex clonal evolution of CLL. They observed a temporal acquisition of mutations and a gradual genomic degeneration with subclones expanding, disappearing, or remaining stable. The presence of clonal evolution was also associated with a shorter OS, which is in line with recent studies.
However, as is often the case, the road from genetic discovery to understanding the biological context, and ultimately to patient therapy, is not always clear-cut and is often plagued by hurdles along the way. In the setting of subclonal mutations in CLL, it is relevant to note that Rasi et al very recently reported no impact of subclonal SF3B1, NOTCH1, and BIRC3 mutations on OS, suggesting that subclonal NOTCH1, SF3B1, and BIRC3 mutations may not provide the same “fitness advantage” as subclonal TP53 mutations.10
Henceforth, given the relative rarity of cases harboring subclonal mutations and the differing clinical impact observed between the 2 aforementioned studies, it is evident that large-scale ultradeep NGS studies must be performed in order to analyze subclonal mutations in CLL and definitively draw conclusions regarding the relative impact of each type of mutation in conjunction with the presence of other genetic events. In other words, we can no longer simply think of mutations as being present or absent, it will be vital to know what proportion of cells harbor a mutation not only to predict outcome but also to optimize therapy. Along these lines, it will be important to address the issue of subclonal mutations in homogeneously treated patients particularly in the era of targeted therapy using B-cell receptor inhibitors (ibrutinib and idelalisib).
As a final remark, while the study by Nadeu et al elegantly demonstrates that as the scope of possibilities increases through technological developments, we are able to ask increasingly sophisticated questions, raising questions regarding (1) how to establish a cutoff value for VAFs, (2) whether this value has to be gene specific, and (3) what the best methodologies are for detecting subclonal mutations. Hence, there is an evident, unmet need to harmonize the methodologies used to detect minor subclones, and rigorous testing is required to establish the technology and minimal requirements for the standardized assessment of such subclones, which will be particularly relevant in the multicenter setting and to ensure clinical benefit before widespread introduction.
Conflict-of-interest disclosure: The authors declare no competing financial interests.