Key Points
The neuronal marker UCH-L1 is induced in, and specifically augments the oncogene-induced transformation of, GCB cells.
High levels of UCHL1 identify patients with GC DLBCL with an increased risk for poor outcomes.
Abstract
Gene expression profiling has identified 2 major subclasses of diffuse large B-cell lymphoma (DLBCL). Cases resembling germinal center (GC) B cells (GCB-DLBCL) generally occur in younger patients, have a distinct molecular pathophysiology, and have improved outcomes compared with those similar to activated post-GC cells (activated B-cell DLBCL). We previously found that the ubiquitin hydrolase UCH-L1 is frequently overexpressed in mature B-cell malignancies and is a potent oncogene in mice. The cause for its overexpression in lymphoma, and whether it impacts the outcome of patients with DLBCL is unknown. Here, we show that UCH-L1 reflects GC lineage in lymphoma and is an oncogenic biomarker of aggressive GCB-DLBCL. We find that UCH-L1 is specifically induced in GC B cells in mice and humans, and that its expression correlates highly with the GCB subtype in DLBCL. We also find that UCH-L1 cooperates with BCL6 in a mouse model of GC B-cell lymphoma, but not with the development of multiple myeloma derived from post-GC cells. Despite the typically good outcomes of GCB-DLBCL, increased UCHL1 identifies a subgroup with early relapses independent of MYC expression, suggesting biological diversity in this subset of disease. Consistent with this, forced Uchl1 overexpression had a substantial impact on gene expression in GC B cells including pathways of cell cycle progression, cell death and proliferation, and DNA replication. These data demonstrate a novel role for UCH-L1 outside of the nervous system and suggest its potential use as a biomarker and therapeutic target in DLBCL.
Introduction
Germinal center (GC) and post-GC-derived B-cell malignancies comprise an important group of cancers that affect children and adults. Diffuse large B-cell lymphoma (DLBCL) can be subclassified based on gene expression signatures into GC B-cell (GCB) or activated B-cell (ABC) types that reflect a GC or post-GC cell of origin, respectively.1 Although associated with superior outcomes,1 many patients with GCB-DLBCL experience relapse of their disease and the overall survival of recurrent DLBCL of any subtype is poor.2,3
Through an unbiased activity screen of deubiquitinating enzymes in a variety of cancers, we uncovered frequent overexpression of the neuroendocrine-specific enzyme UCH-L1 in mature B-cell cancers including Burkitt lymphoma and DLBCL.4,5 We subsequently found transgenic Uchl1 drives the development of spontaneous lymphoma in mice, demonstrating its oncogenic activity.5 Mechanistically, UCH-L1 plays a novel role in regulating mammalian target of rapamycin (mTOR)-AKT signaling, a pathway important in GCB and lymphoma development.6,7 Despite its frequent overexpression, there are no chromosome translocations, copy number alterations, or point mutations known to affect UCH-L1 levels. Here, we report that UCH-L1 expression is specifically induced in GC B cells, and its expression reflects GC identity in lymphoma. Forced expression of UCH-L1 promotes oncogenic gene expression patterns in GC B cells and accelerates lymphomagenesis driven by the GC regulator and oncogene BCL6. Importantly, we find that increased UCHL1 identifies patients with a poor prognosis specifically in GCB-DLBCL. We conclude that UCH-L1 expression in lymphoma reflects GCB gene expression patterns in lymphoma and may represent a novel prognostic marker and therapeutic target in this disease.
Methods
Reagents and general procedures
Antibodies include BCL6 (Santa Cruz Biotechnology, Dallas, TX, and Cell Signaling Technology, Danvers, MA), IRF4, Histone H2B, Tubulin, p-AKTS473, AKT (Cell Signaling Technology), BCL2 (R&D Systems, Minneapolis, MN), B220, GL7, IgG1, and CD138 (BD Pharmingen, San Jose, CA), CD23, and UCH-L1 (Thermo Scientific, Waltham, MA). Biotin-conjugated secondary antibodies were from Vector Laboratories (Burlingame, CA). Cells were cultured in complete RPMI 1640 (high glucose with pyruvate and glutamine) supplemented with 10% stem cell qualified fetal bovine serum (Gemini Bio-Products, West Sacramento, CA). Lentivirus-encoded short-hairpin RNAs (shRNAs) were generated and used as described.5,8 Cell viability was monitored using the MTS (3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium) assay as described.5,8 Flow cytometry was performed and analyzed with an Accuri C6 cytometer (Accuri Cytometers Inc, Ann Arbor, MI), using BD Accuri C6 software version 1.0.264.21. Quantitative real-time polymerase chain reaction (PCR) was performed using TaqMan probes for mouse Uchl1 normalized to Gapdh (Applied Biosystems). Fold-change was calculated with the Δ-Δ cycle threshold method. Tumor clonality was determined as described.9,10
Mice, immunizations, isolation of GCBs, and antigen-specific immunity
Uchl1Tg,5 IμHABCL6,10 and Vk*MYC mice11 were housed in barrier conditions in accordance with protocols approved by the institutional animal care and use committee of the Mayo Clinic. GCB and non-GCB isolation was carried out as described12 and purity confirmed by flow cytometry (BD Accuri C6; BD Biosciences, San Jose, CA). To monitor antigen-specific immune responses, mice (3-4 months of age) were immunized with 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to keyhole limpet hemocyanin (NP-KLH; Biosearch Technologies, Petaluma, CA) in Freund complete adjuvant (Sigma). Enzyme-linked immunosorbent assay (ELISA) plates coated with NP(25)-bovine serum albumin (Biosearch) were incubated with a 1/5000 dilution of serum samples collected on days 0, 7, and 14. Captured molecules where detected by horseradish peroxidase–conjugated anti-mouse immunoglobulin G1 (IgG1).
Histology and immunohistochemistry
Formalin-fixed paraffin-embedded tissues were used for histology, immunohistochemistry, and immunofluorescence. Antigen retrieval was performed with the Vector Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA). Slides were developed by diaminobenzidine (Vector Laboratories) or fluorescent-labeled secondary antibodies (R&D Systems). The terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was performed using the TACS In situ Apoptosis Detection kit (Trevigen Inc, Gaithersburg, MD). The tissue microarray was created as part of the Mayo Clinic Lymphoma Specialized Program of Research Excellence (SPORE) Molecular Epidemiology Resource, a prospective cohort of over 5000 lymphoma patients. All research involving human samples was reviewed and approved by the Mayo Clinic Institutional Review Board.
Analysis of gene expression data and statistics
RNA was isolated using RNeasy (Qiagen) from GCB from IμHABCL6 or Uchl1Tg/IμHABCL6 mice (n = 3 each); gene expression was measured on an Affymetrix Mouse 430 2.0 array through the Mayo Clinic Gene Expression Core. All samples were preprocessed and normalized using Partek software. Robust multiarray average normalization was applied to the samples and the expression signals were converted to log2 scale for downstream analysis. The data were analyzed for quality control using principal component analysis and hierarchal clustering from Partek. The standard analysis of variance model from Partek was applied to calculate the differentially expressed genes for each of the comparisons. Gene expression and clinical annotations from GSE2350,13 GSE4475,14 GSE31312,15 GSE3613316 (human), and GSE38696, GSE3869717 GSE39403,18 GSE3830419 (mouse) were extracted from the NCBI Gene Expression Omnibus (GEO) database and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). Normalized log2 transformed gene expression data were downloaded from the R2 platform to a Microsoft Excel spreadsheet for additional analysis. All statistics were performed using GraphPad Prism software with the exception of the univariate and multivariate analyses, which were performed using JMP (SAS).
Results
UCH-L1 overexpression is characteristic of Burkitt lymphoma and GCB-DLBCL
To understand its impact on aggressive B-cell lymphomas, we developed a profile of UCH-L1–expressing lymphomas. We classified 215 cases of B-cell lymphoma as either UCHL1 LO (0%-79%) or UCHL1 HI (80%-100%) based on gene expression profiling.14 Comparing the UCHL1 HI cases with the overall cohort, there was a highly significant difference in the morphologic classification, with UCHL1 HI cases more likely to have a Burkitt or atypical Burkitt histology (Table 1). Of those classified as DLBCL, there was a significant enrichment (P = .007) of cases with the GCB signature. Within the UCHL1 HI cases, there was also a significant increase in cases molecularly classified as Burkitt lymphoma (mBL). Consistent with this, UCHL1 HI cases were significantly more likely to carry the immunoglobulin-MYC translocation (Table 1). There was also a significant difference in immunohistochemical patterns, as UCHL1 HI cases were more likely to be BCL2-negative, BCL6-positive, and less likely to have a break in the BCL6 locus (Table 1).
Characteristics of UCHL1-expressing lymphomas
| Characteristic . | All . | UCHL1 HI (80%-100%) . | P . |
|---|---|---|---|
| No. (%) . | |||
| Total | 215 | 43 | |
| Morphologic Dx | <.0001 | ||
| DLBCL | 162 (75) | 15 (35) | |
| BL | 8 (4) | 6 (14) | |
| Atypical BL | 28 (13) | 17 (40) | |
| Unclassified aggressive mature B-NHL | 18 (8) | 5 (11) | |
| GEP | .0012 | ||
| ABC | 55 (26) | 3 (7) | |
| GCB | 117 (54) | 35 (81) | |
| Unclassified | 43 (20) | 5 (12) | |
| Mol. Dx. | <.0001 | ||
| mBL | 44 (21) | 32 (74) | |
| Non-mBL | 125 (58) | 8 (19) | |
| Intermediate | 46 (21) | 3 (7) | |
| BCL2 IHC | <.0001 | ||
| Absent | 61 (29) | 26 (60) | |
| Present | 149 (69) | 15 (35) | |
| NA | 5 (2) | 2 (5) | |
| BCL6 IHC | .0435 | ||
| Absent | 29 (13) | 2 (5) | |
| Present | 165 (77) | 40 (93) | |
| NA | 21 (10) | 1 (2) | |
| BCL6 break | .0405 | ||
| Absent | 173 (80) | 41 (95) | |
| Present | 36 (17) | 2 (5) | |
| NA | 6 (3) | 0 (0) | |
| IgH-BCL2 fusion | NS | ||
| Absent | 188 (87) | 38 (88) | |
| Present | 25 (12) | 5 (12) | |
| NA | 2 (1) | 0 (0) | |
| MYC status | <.0001 | ||
| MYC neg. | 139 (65) | 10 (24) | |
| Ig-MYC | 57 (26) | 31 (72) | |
| Non-Ig-MYC | 15 (7) | 1 (2) | |
| NA | 4 (2) | 1 (2) |
| Characteristic . | All . | UCHL1 HI (80%-100%) . | P . |
|---|---|---|---|
| No. (%) . | |||
| Total | 215 | 43 | |
| Morphologic Dx | <.0001 | ||
| DLBCL | 162 (75) | 15 (35) | |
| BL | 8 (4) | 6 (14) | |
| Atypical BL | 28 (13) | 17 (40) | |
| Unclassified aggressive mature B-NHL | 18 (8) | 5 (11) | |
| GEP | .0012 | ||
| ABC | 55 (26) | 3 (7) | |
| GCB | 117 (54) | 35 (81) | |
| Unclassified | 43 (20) | 5 (12) | |
| Mol. Dx. | <.0001 | ||
| mBL | 44 (21) | 32 (74) | |
| Non-mBL | 125 (58) | 8 (19) | |
| Intermediate | 46 (21) | 3 (7) | |
| BCL2 IHC | <.0001 | ||
| Absent | 61 (29) | 26 (60) | |
| Present | 149 (69) | 15 (35) | |
| NA | 5 (2) | 2 (5) | |
| BCL6 IHC | .0435 | ||
| Absent | 29 (13) | 2 (5) | |
| Present | 165 (77) | 40 (93) | |
| NA | 21 (10) | 1 (2) | |
| BCL6 break | .0405 | ||
| Absent | 173 (80) | 41 (95) | |
| Present | 36 (17) | 2 (5) | |
| NA | 6 (3) | 0 (0) | |
| IgH-BCL2 fusion | NS | ||
| Absent | 188 (87) | 38 (88) | |
| Present | 25 (12) | 5 (12) | |
| NA | 2 (1) | 0 (0) | |
| MYC status | <.0001 | ||
| MYC neg. | 139 (65) | 10 (24) | |
| Ig-MYC | 57 (26) | 31 (72) | |
| Non-Ig-MYC | 15 (7) | 1 (2) | |
| NA | 4 (2) | 1 (2) |
Data extracted from GEO dataset GSE4475.
B-NHL, B-cell non-Hodgkin's lymphoma; BL, Burkitt lymphoma; Dx, diagnosis; IHC, immunohistochemistry; Mol. Dx., molecular diagnosis; NA, not applicable; neg., negative; NS, not significant.
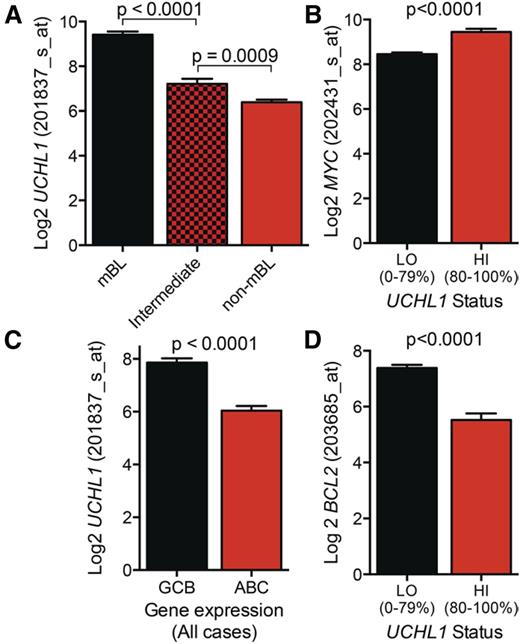
To complement these data, we examined the gene expression profiles (GEPs) of these cases. Cases classified as mBL by GEP had a significantly higher level of UCHL1 compared with either non-mBL or intermediate cases (Figure 1A). As expected based on the enrichment of mBL and immunoglobulin-MYC translocations, UCHL1 HI tumors had significantly increased MYC expression (Figure 1B). Cases classified as GCB also had a significant increase in UCHL1 compared with ABC (Figure 1C). As predicted by immunohistochemistry, there was a significantly lower mean BCL2 level by gene expression profiling in those cases with high UCHL1 (Figure 1D). To further verify the expression of UCH-L1 in DLBCL, we performed immunohistochemistry on a tissue microarray containing 250 cases of DLBCL (149 GCB). Of these cases, a total of 151 (60%) had at least some immunoreactivity for UCH-L1 and 33 (13%) had strong immunoreactivity (supplemental Figure 1A-C, available on the Blood Web site). In 104 cases (42% of the total cases; 69% of those with any immunoreactivity), the percentage of immunopositivity was <50% of the tumor cells. There was a significant increase in the number of positive cases in the GCB group (68% vs 53%; P = .05). These data demonstrate that UCH-L1 protein is detectable by immunohistochemistry in DLBCL. Taken together, we conclude that B-cell lymphomas that express high levels of UCH-L1 are likely to have gene expression and immunophenotypic characteristics of GC lineage.
UCHL1 expression is highest in GCB-derived lymphomas. (A,C) The expression of UCHL1 as reflected in RNA microarray data are shown for a series of 215 cases of mature B-cell lymphoma classified based on molecular classification as either mBL, non-mBL, or intermediate (A) or cell-of-origin gene expression classification including both mBL and non-mBL cases (C). (B,D) Expression of MYC (B) or BCL2 (D) in cases from panel A classified based on the level of UCHL1 as shown. Data were extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). All P values were calculated using the Student t test.
UCHL1 expression is highest in GCB-derived lymphomas. (A,C) The expression of UCHL1 as reflected in RNA microarray data are shown for a series of 215 cases of mature B-cell lymphoma classified based on molecular classification as either mBL, non-mBL, or intermediate (A) or cell-of-origin gene expression classification including both mBL and non-mBL cases (C). (B,D) Expression of MYC (B) or BCL2 (D) in cases from panel A classified based on the level of UCHL1 as shown. Data were extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). All P values were calculated using the Student t test.
UCH-L1 is expressed in GCBs
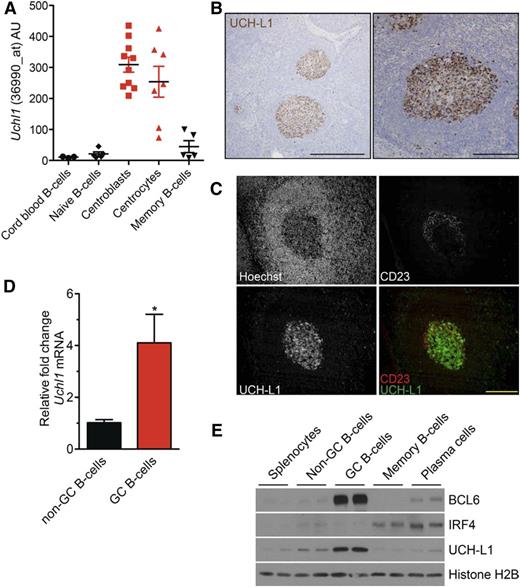
The mechanisms for increased UCH-L1 expression in lymphoma are not clear. According to the COSMIC database (http://cancer.sanger.ac.uk/cosmic), there are no reported mutations or chromosome alterations that involve the UCHL1 locus, suggesting direct transcriptional regulation. Although UCH-L1 is widely regarded as a neuroendocrine-specific enzyme,20 we reasoned that whole-tissue GEP might easily miss its expression in the relatively small population of GCB. We therefore examined data from purified human B-cell populations.13 Although UCHL1 levels were very low in naive and memory B cells, its expression sharply increased in centroblast and centrocyte GCB populations (Figure 2A). Strikingly, there is a very similar pattern of expression in cancer cell lines derived from these stages of development (supplemental Figure 2).5,16 We next performed immunohistochemistry on human reactive lymph nodes and found a striking localization of UCH-L1 protein to GCs (Figure 2B). There was no evidence of polarized expression between the light and dark zones (Figure 2C). Consistent with this, UCHL1 messenger RNA (mRNA) was not substantially different in these populations in either human or mouse (supplemental Figure 3).17 We also found high levels of Uchl1 mRNA and protein in purified GCB from mice (Figure 2D-E), and the strong induction of UCH-L1 expression in B cells undergoing an in vitro class switch recombination assay (supplemental Figure 4A-B). The expression of UCH-L1 in GC B cells is not induced by BCL6, as purified GCB from mice with deregulated expression of BCL6 (IμHABCL6)10 do not have higher levels of UCH-L1 (supplemental Figure 5A). In previous studies, neither BCL6 depletion, nor the overexpression of degradation-deficient BCL6, changed UCHL1 levels.21,22 Published chromatin immunoprecipitation–sequencing (ChIP-seq) data did not find BCL6 occupancy of the UCHL1 promoter.23 We also examined whether UCH-L1 was induced by MYC. Two independent studies using Myc–green fluorescent protein (GFP) reporter mouse strains recently examined the role of MYC in GCB development.18,19 In these reports, gene expression in Myc-GFP-negative or -positive purified GCB showed no changes in Uchl1 transcript level18,19 (supplemental Figure 5B), providing compelling evidence that MYC does not induce UCH-L1 expression. Taken together, these data demonstrate that UCH-L1 is specifically induced in GCB, and suggest that its expression in B-cell lymphoma reflects its expression in the cell or origin.
UCH-L1 is specifically induced in GCBs. (A) The expression of UCHL1 as reflected in RNA microarray data are shown for the indicated purified human B-cell subsets. Data extracted from GSE2350.13 (B) Formalin-fixed paraffin-embedded human reactive lymph node specimens were stained for UCH-L1 (brown). Bar: 500 μm (left), 200 μm (right). (C) Formalin-fixed paraffin-embedded human tonsil sections were stained with the indicated. Bar: 200 μm. (D) Quantitative real-time PCR for murine Uchl1 was performed on complementary DNA generated from GC or non-GCBs purified from wild-type mice (n = 3 each); *P < .05. (E) Extracts were prepared from the indicated purified B-cell subsets (n = 2 each), and samples were immunoblotted for the indicated proteins. Histone H2B is included as a loading control. Microscopy images were obtained with an Olympus AX70 microscope with a DP71 camera.
UCH-L1 is specifically induced in GCBs. (A) The expression of UCHL1 as reflected in RNA microarray data are shown for the indicated purified human B-cell subsets. Data extracted from GSE2350.13 (B) Formalin-fixed paraffin-embedded human reactive lymph node specimens were stained for UCH-L1 (brown). Bar: 500 μm (left), 200 μm (right). (C) Formalin-fixed paraffin-embedded human tonsil sections were stained with the indicated. Bar: 200 μm. (D) Quantitative real-time PCR for murine Uchl1 was performed on complementary DNA generated from GC or non-GCBs purified from wild-type mice (n = 3 each); *P < .05. (E) Extracts were prepared from the indicated purified B-cell subsets (n = 2 each), and samples were immunoblotted for the indicated proteins. Histone H2B is included as a loading control. Microscopy images were obtained with an Olympus AX70 microscope with a DP71 camera.
UCH-L1 accelerates BCL6-driven B-cell lymphomagenesis
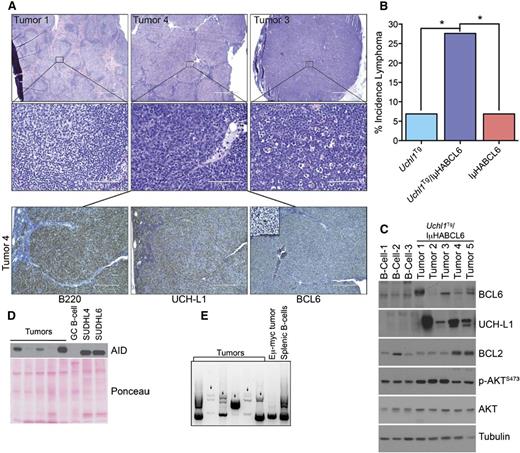
To further examine its impact on GC-derived lymphomagenesis, we crossed transgenic mice that express UCH-L1 in a broad tissue distribution (Uchl1Tg)5 with mice carrying deregulated BCL6.10 In this model, HA-tagged BCL6 was inserted into the immunoglobulin heavy chain enhancer (IμHABCL6). We generated cohorts of Uchl1, IμHABCL6, and Uchl1/IμHABCL6 double mutant mice and analyzed them for lymphomas. As expected based on their young age, there was a low rate of B-cell lymphoma in either the Uchl1 or IμHABCL6 strains (Figure 3A-B). However, we observed a significant increase in lymphomagenesis in the double mutant mice. The tumors were of B-cell lineage, and exhibited a range in histology spanning from follicular-type lymphomas to DLBCLs (Figure 3C). As has been reported previously, BCL6 expression is difficult to detect in some tumors using immunohistochemistry, though BCL6 and UCH-L1 protein was seen in all cases by immunoblotting (Figure 3C). Of the 5 tumors analyzed, 3 had increased levels of p-AKTS473 and 2 had increased BCL2. The tumors also expressed substantial amounts of activation-induced cytidine deaminase (Figure 3D). Clonality of the tumors was confirmed by immunoglobulin PCR (Figure 3E). Taken together, these data demonstrate that UCH-L1 cooperates with BCL6 in B-cell lymphomagenesis.
Transgenic UCH-L1 synergizes with deregulated BCL6 in the development of B-cell lymphoma. (A) Representative histology of lymphomas observed in Uchl1Tg/IμHABCL6 mice. Formalin-fixed paraffin-embedded sections were stained with hematoxylin and eosin (top 2 rows) or immunohistochemistry (bottom row) with antibodies against the indicated antigens. Bar: 1000 μm (top row), 100 μm (middle row), 400 μm (bottom row), 100 μm (inset, bottom row). Microscopy images were obtained with an Olympus AX70 microscope with a DP71 camera. (B) The incidence of lymphomas is shown for the indicated mouse strains. *P < .05 as determined with the χ2 test; N = 29 mice for each genotype. (C-D) Immunoblots were performed on extracts from lymphomas as in panel A. Comparison is made with purified B cells (C), purified GCBs (D) from the spleens of wild-type littermates, or the indicated GCB-DLBCL cell lines (D). (E) Genomic DNA was extracted from the indicated samples and subjected to PCR amplification of immunoglobulin variable regions. The arrows denote unique monoclonal bands not seen in the polyclonal B cells.
Transgenic UCH-L1 synergizes with deregulated BCL6 in the development of B-cell lymphoma. (A) Representative histology of lymphomas observed in Uchl1Tg/IμHABCL6 mice. Formalin-fixed paraffin-embedded sections were stained with hematoxylin and eosin (top 2 rows) or immunohistochemistry (bottom row) with antibodies against the indicated antigens. Bar: 1000 μm (top row), 100 μm (middle row), 400 μm (bottom row), 100 μm (inset, bottom row). Microscopy images were obtained with an Olympus AX70 microscope with a DP71 camera. (B) The incidence of lymphomas is shown for the indicated mouse strains. *P < .05 as determined with the χ2 test; N = 29 mice for each genotype. (C-D) Immunoblots were performed on extracts from lymphomas as in panel A. Comparison is made with purified B cells (C), purified GCBs (D) from the spleens of wild-type littermates, or the indicated GCB-DLBCL cell lines (D). (E) Genomic DNA was extracted from the indicated samples and subjected to PCR amplification of immunoglobulin variable regions. The arrows denote unique monoclonal bands not seen in the polyclonal B cells.
We next tested whether UCH-L1 also accelerates the development of myeloma. To address this, we mated Uchl1Tg mice with a transgenic model in which the human MYC oncogene is driven by the mouse κ light-chain locus (Vk*MYC).11 With age, these animals develop multiple myeloma with biological features that closely resemble human disease. Consistent with prior reports, Vk*MYC mice develop m-spikes prior to 20 weeks of age with the incidence reaching 100% by 80 weeks, with nests of CD138+ cells in the bone marrow of affected mice consistent with transformed plasma cells (supplemental Figure 6A-B). The onset and progression of m-spike development was unchanged in Vk*MYC/Uchl1Tg double mutant mice. We additionally found no UCH-L1–associated changes in the numbers of marrow plasma cells, serum IgG, or blood hemoglobin (a sign of end organ dysfunction) in these mice (supplemental Figure 6C-E). We conclude that UCH-L1 specifically cooperates with the generation of cancers derived from GCBs.
UCH-L1 is a prognostic marker in GC-derived B-cell lymphomas
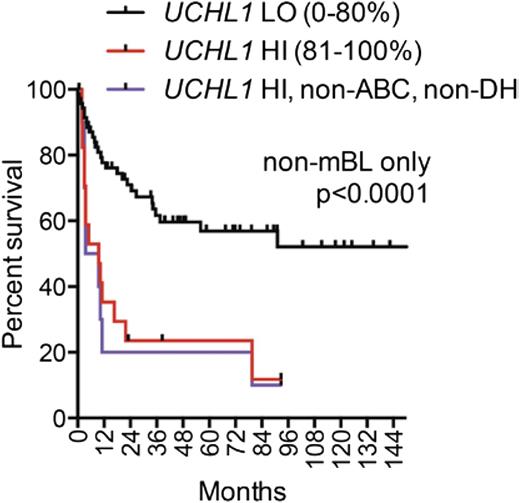
To assess whether UCH-L1 expression has an impact on outcomes, we turned to a clinically annotated gene expression data set14 of 155 cases with paired GEP and outcome data consisting of 30 cases of mBL, 92 non-mBL, and 33 intermediate defined by GEP. Within the mBL group, most samples have high levels of UCHL1, as seen in Table 1. There was no difference in outcome in mBL cases with low (n = 7) vs those with high levels (n = 23) (data not shown). We next analyzed the outcomes in those patients with non-mBL, with UCHL1 HI defined as the top 20% of the expression range within this subgroup (n = 17). There was a highly significant and substantial reduction in survival in the UCHL1 HI cohort compared with the remaining patients (Figure 4). Two factors known to have a powerful impact on outcomes include ABC GEP and so called “double-hit” lymphoma, defined as the combined occurrence (any 2) of MYC, BCL2, or BCL6 rearrangements.24 Of the patients with increased UCHL1, there were 4 ABC subtypes and 3 double hit cases. If these 7 patients are removed, the survival remains severely inferior, indicating that the negative impact of high UCHL1 expression is independent of these known poor prognostic factors (Figure 4). Although the patient cohort was not uniformly treated, most (75%) received a “CHOP-like” regimen, and 16% received rituximab.
High levels of UCHL1 mark patients with non-mBL at high risk for relapse. Overall survival is plotted using the Kaplan-Meier method for 92 patients with non-mBL stratified based on the level of UCHL1 mRNA as shown. Those with ABC gene expression profile and double hit (DH) lymphoma (as defined by reported fluorescence in situ hybridization studies for MYC, BCL2, and BCL6) were excluded as shown. Data were extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). P value was determined using the Mantel-Cox log-rank test.
High levels of UCHL1 mark patients with non-mBL at high risk for relapse. Overall survival is plotted using the Kaplan-Meier method for 92 patients with non-mBL stratified based on the level of UCHL1 mRNA as shown. Those with ABC gene expression profile and double hit (DH) lymphoma (as defined by reported fluorescence in situ hybridization studies for MYC, BCL2, and BCL6) were excluded as shown. Data were extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). P value was determined using the Mantel-Cox log-rank test.
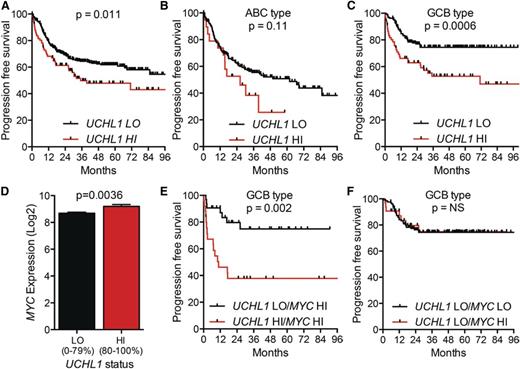
To examine impact of UCH-L1 in patients treated with modern therapy, we turned to the International DLBCL Rituximab-CHOP Consortium Program, a data set containing 470 patients with DLBCL uniformly treated with rituximab with cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) and having clinical data and GEP.15 Examining the cohort without regard to cell of origin revealed that increased levels of UCHL1 (80%-100%) were associated with a significant reduction in progression-free survival (PFS) (Figure 5A). Comparing the UCHL1 LO and HI groups, there was no difference in clinical stage or the proportion of patients with high International Prognostic Index (IPI) scores (supplemental Figure 7A-B). We again observed a significant increase in UCHL1 HI cases classified as GCB subtype (supplemental Figure 7C). Subgroup analysis revealed a highly significant negative impact of increased UCHL1 on PFS in the GCB subgroup, but not in the ABC group (Figure 5B-C). As seen in the other cohort, there was an increase in MYC in the UCHL1 HI cases (Figure 5D). Strikingly, patients with increased MYC (75%-100% expression range) and UCHL1 had a far worse outcome compared with increased MYC alone (Figure 5E). Additionally, the negative impact of high MYC on outcome was entirely lost in cases with low UCHL1 (Figure 5F). To further explore the relationship between UCHL1 and MYC, we conducted univariate and multivariate analysis. For the univariate analysis, we also analyzed the impact of EZH2 levels as high levels were recently found to be favorable in GCB-DLBCL.25 As expected, high levels of MYC, UCHL1, and low levels of EZH2 (<85% maximum) all had a significant impact on PFS in the univariate analysis, and all continued to be significant on multivariate analysis (Tables 2-3). We conclude that UCHL1 and MYC have an additive impact on outcome, suggesting independent mechanisms in the pathophysiology of GCB-DLBCL.
High levels of UCHL1 predict poor outcomes in patients with GCB-DLBCL treated with R-CHOP. (A-C) PFS for all patients with paired gene expression and survival data from the International DLBCL Rituximab-CHOP Consortium Program study (n = 470) (A), those with ABC-DLBCL (n = 199) (B), and GCB-DLBCL (n = 227) (C). UCHL1 was stratified with LO (=0%-80%) HI (=80%-100%) based on the entire cohort. P values were determined using the Mantel-Cox log-rank test. (D) The level of MYC mRNA is shown for the entire cohort from panel A including those lacking survival data (n = 498). UCHL1 stratification as in panels A-C. P value was determined using the Student t test. (E-F) PFS of patients with GCB-DLBCL (n = 227) with paired gene expression and survival data based on the stratification of UCHL1 and MYC. UCHL1 was stratified as above. MYC was stratified with LO (=0%-75%) and HI (=75%-100%). Data extracted from GSE31312 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl).
High levels of UCHL1 predict poor outcomes in patients with GCB-DLBCL treated with R-CHOP. (A-C) PFS for all patients with paired gene expression and survival data from the International DLBCL Rituximab-CHOP Consortium Program study (n = 470) (A), those with ABC-DLBCL (n = 199) (B), and GCB-DLBCL (n = 227) (C). UCHL1 was stratified with LO (=0%-80%) HI (=80%-100%) based on the entire cohort. P values were determined using the Mantel-Cox log-rank test. (D) The level of MYC mRNA is shown for the entire cohort from panel A including those lacking survival data (n = 498). UCHL1 stratification as in panels A-C. P value was determined using the Student t test. (E-F) PFS of patients with GCB-DLBCL (n = 227) with paired gene expression and survival data based on the stratification of UCHL1 and MYC. UCHL1 was stratified as above. MYC was stratified with LO (=0%-75%) and HI (=75%-100%). Data extracted from GSE31312 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl).
Univariate analysis of prognostic factors
| Factor . | RR . | 95% CI . | P . |
|---|---|---|---|
| UCHL1 >80% | 2.26 | 1.39-3.64 | .0012 |
| MYC >85% | 2.21 | 1.18-3.87 | .0148 |
| EZH2 <85% | 2.6 | 1.16-7.43 | .0184 |
| Factor . | RR . | 95% CI . | P . |
|---|---|---|---|
| UCHL1 >80% | 2.26 | 1.39-3.64 | .0012 |
| MYC >85% | 2.21 | 1.18-3.87 | .0148 |
| EZH2 <85% | 2.6 | 1.16-7.43 | .0184 |
N = 227 cases of GCB-DLBCL uniformly treated with R-CHOP.
CI, confidence interval; RR, relative risk.
Multivariate proportional hazard analysis of prognostic factors
| Factor . | RR . | 95% CI . | P . |
|---|---|---|---|
| UCHL1 >80% | 2.4 | 1.46-3.91 | .0007 |
| MYC >85% | 1.89 | 1.00-3.35 | .0495 |
| EZH2 <85% | 3.29 | 1.45-9.48 | .0028 |
| Factor . | RR . | 95% CI . | P . |
|---|---|---|---|
| UCHL1 >80% | 2.4 | 1.46-3.91 | .0007 |
| MYC >85% | 1.89 | 1.00-3.35 | .0495 |
| EZH2 <85% | 3.29 | 1.45-9.48 | .0028 |
N = 227 cases of GCB-DLBCL uniformly treated with R-CHOP.
Abbreviations explained in Table 2.
UCH-L1 promotes mTORC2 activity, AKT phosphorylation, and cell survival in DLBCL
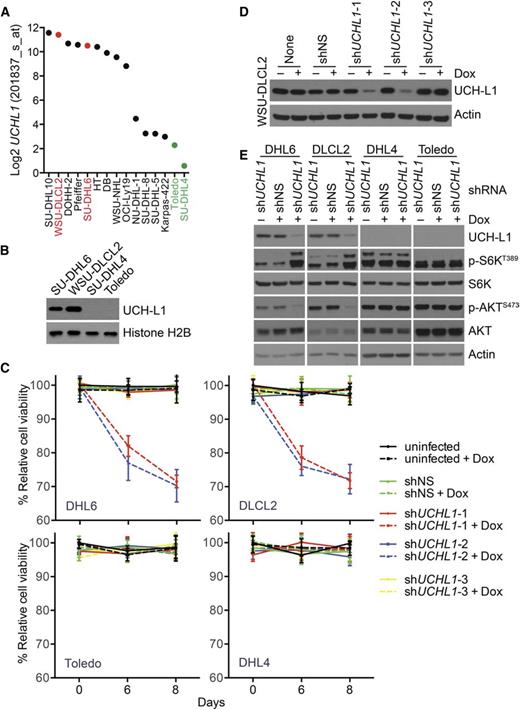
We previously observed that UCH-L1 affects the stability of mTOR complex 1 (mTORC1) resulting in increased formation of mTORC2 and enhanced phosphorylation of AKTS473.5,8 To determine whether a similar mechanism operates in DLBCL cells, we examined the expression of UCHL1 in the cancer cell line encyclopedia (CCLE; http://www.broadinstitute.org/ccle/home) and identified a set of GCB-DLBCL cell lines with varied expression of UCHL1, the pattern of which we confirmed by immunoblotting (Figure 6A-B). We used a set of doxycycline (Dox)-inducible shRNA constructs to determine the impact of UCH-L1 depletion in these cells. Compared with untransduced cells and a non-silencing control, 2 of the 3 UCHL1-targeting shRNAs reduced UCH-L1 protein levels (Figure 6D). The 2 hairpins that reduce UCH-L1 protein selectively impair the proliferation of the GCB-DLBCL cell lines with high levels of UCH-L1 (Figure 6C-D). This indicates that, when expressed, UCH-L1 provides an oncogenic signal that promotes the proliferation of DLBCL cells. To better understand this signal, we depleted UCH-L1 and examined the impact on mTOR-AKT signaling. Compared with untransduced cells and a nonsilencing control, depletion of UCH-L1 led to increased phosphorylation of p70S6 kinase (p-S6KT389) and reduced phosphorylation of AKTS473, indicative of increased mTORC1 and reduced mTORC2 activity, respectively (Figure 6E). One of the critical targets for AKT in promoting survival is FOXO1. Phosphorylation of FOXO1 leads to its cytoplasmic sequestration and a reduction in its ability to promote the transcription of its targets. We therefore examined the level of UCHL1 and that of a list of well-characterized FOXO1 targets in mature B-cell lymphomas. Of the 9 FOXO1 targets examined, all but 1 was significantly (P < .05) correlated with the level of UCHL1 (supplemental Figure 8A-B). Of these, 6 of the 9 FOXO1 targets were inversely proportional to UCHL1, consistent with decreased FOXO1 activity (supplemental Figure 8B-D). Taken together, these data lead us to conclude that UCH-L1 promotes AKT signaling and cell survival in DLBCL.
UCH-L1 promotes AKT phosphorylation and cell survival in DLBCL. (A) The expression of UCHL1 is shown for a series of GCB-DLBCL cell lines. The cell lines used in this study are shown in red (UCHL1 HI) and green (UCHL1 LO). Data extracted from the CCLE (GSE36133) and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). (B) Immunoblot analysis of the cell lines from panel A. Histone H2B is included as a loading control. (C-D) The cell lines from panel A were transduced with lentivirus’ encoding the indicated Dox-inducible shRNA constructs. Cell viability (C) was monitored using the MTS assay. The level of UCH-L1 (D) is shown for the WSU-CLCL2 cell line. shNS, control nonsilencing; shUCHL1, n = 3 independent UCHL1 targeting shRNA constructs. Similar results were obtained in SU-DHL6 cells. (E) The indicated cell lines were transduced with the indicated shRNA constructs as in panels C and D and the resulting extracts were analyzed with antibodies against the indicated proteins. Dox was included where indicated.
UCH-L1 promotes AKT phosphorylation and cell survival in DLBCL. (A) The expression of UCHL1 is shown for a series of GCB-DLBCL cell lines. The cell lines used in this study are shown in red (UCHL1 HI) and green (UCHL1 LO). Data extracted from the CCLE (GSE36133) and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). (B) Immunoblot analysis of the cell lines from panel A. Histone H2B is included as a loading control. (C-D) The cell lines from panel A were transduced with lentivirus’ encoding the indicated Dox-inducible shRNA constructs. Cell viability (C) was monitored using the MTS assay. The level of UCH-L1 (D) is shown for the WSU-CLCL2 cell line. shNS, control nonsilencing; shUCHL1, n = 3 independent UCHL1 targeting shRNA constructs. Similar results were obtained in SU-DHL6 cells. (E) The indicated cell lines were transduced with the indicated shRNA constructs as in panels C and D and the resulting extracts were analyzed with antibodies against the indicated proteins. Dox was included where indicated.
UCH-L1 alters gene expression and antigen-specific immunity in premalignant GCBs
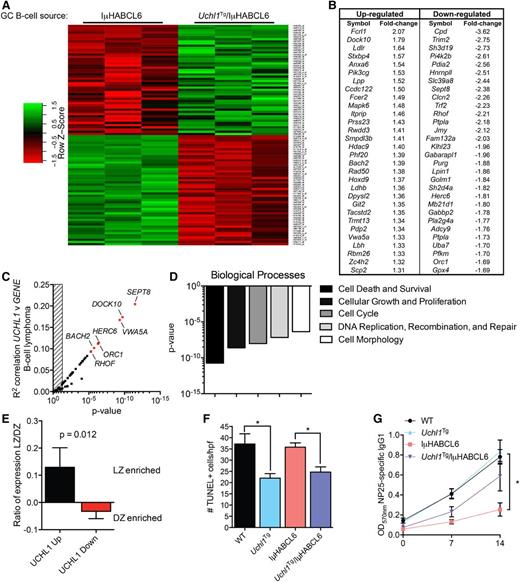
To further gauge the impact of UCH-L1 on GC-derived lymphomagenesis, we performed gene expression profiling on premalignant GCBs purified from young (3-6 months) IμHABCL6 and Uchl1/IμHABCL6 mice (n = 3 each genotype). Transgenic UCH-L1 resulted in the differential expression (P < .05) of 1884 probe sets composed of 951 upregulated and 933 downregulated genes (Figure 7A-B). Of these, there were 320 or 59 probe sets changed at least 1.5- or 2-fold (P < .05), respectively. We next examined the correlation of the top 60 altered genes with UCHL1 levels in mature B-cell lymphomas (n = 215). Of these, 41 of 60 showed a correlation with UCHL1 (Figure 7C). To gain insight into the nature of these changes, we performed Ingenuity Pathway Analysis (IPA) on those significantly changed probe sets (Figure 7D). There was a strong alteration in the biological processes of cell death and survival, cell growth and proliferation, cell cycle, DNA replication, recombination, and repair, and cell morphology.
Transgenic UCH-L1 leads to altered gene expression in GCBs. (A-B) Gene expression profiling was performed using RNA extracted from GCBs from mice of the indicated genotypes (n = 3 each). (A) A heat map represents the relative expression of the 100 most discriminatory genes. (B) The 60 most altered genes (30 up, 30 down; P < .01) are shown with the mean fold-change indicated. (C) The list of genes from panel B were correlated with UCHL1 levels in 215 mature B-cell lymphomas extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). The graph represents the Pearson correlation R2 value and the corresponding P value for each gene. The hatched area on the left represents a P > .05. Genes with the most significant correlation are indicated in red. (D) IPA was performed using the list of differentially expressed genes (P < .05) from panel A. The graph represents the P values of the top 5 altered biological processes. (E) The expression ratio (mean ± SE) of the list of top upregulated and downregulated genes from panel B in purified mouse light zone (LZ) or dark zone (DZ) cells (GSE3869617 ) is shown. The P value was calculated with the Student t test. (F) Formalin-fixed paraffin-embedded spleen sections from mice of the indicated genotypes were stained using the TUNEL assay. The graph represents the mean ± SE number of TUNEL-positive cells from 10 randomly selected high-power field (hpf) images from each of 3 mice per genotype (total of 30 hpf per genotype). P values were calculated with the Student t test. (G) Mice of the indicated genotypes (n = 6 each) were immunized with NP-CGG and the level of NP-specific IgG1 was determined by ELISA on the indicated days. *P < .05 (Student t test) for the comparison between IμHABCL6 and any of the other genotypes.
Transgenic UCH-L1 leads to altered gene expression in GCBs. (A-B) Gene expression profiling was performed using RNA extracted from GCBs from mice of the indicated genotypes (n = 3 each). (A) A heat map represents the relative expression of the 100 most discriminatory genes. (B) The 60 most altered genes (30 up, 30 down; P < .01) are shown with the mean fold-change indicated. (C) The list of genes from panel B were correlated with UCHL1 levels in 215 mature B-cell lymphomas extracted from GSE447514 and analyzed using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). The graph represents the Pearson correlation R2 value and the corresponding P value for each gene. The hatched area on the left represents a P > .05. Genes with the most significant correlation are indicated in red. (D) IPA was performed using the list of differentially expressed genes (P < .05) from panel A. The graph represents the P values of the top 5 altered biological processes. (E) The expression ratio (mean ± SE) of the list of top upregulated and downregulated genes from panel B in purified mouse light zone (LZ) or dark zone (DZ) cells (GSE3869617 ) is shown. The P value was calculated with the Student t test. (F) Formalin-fixed paraffin-embedded spleen sections from mice of the indicated genotypes were stained using the TUNEL assay. The graph represents the mean ± SE number of TUNEL-positive cells from 10 randomly selected high-power field (hpf) images from each of 3 mice per genotype (total of 30 hpf per genotype). P values were calculated with the Student t test. (G) Mice of the indicated genotypes (n = 6 each) were immunized with NP-CGG and the level of NP-specific IgG1 was determined by ELISA on the indicated days. *P < .05 (Student t test) for the comparison between IμHABCL6 and any of the other genotypes.
To relate these expression changes to GC biology, we compared the altered genes to a list of genes with polarized expression between the light zone and dark zone GC cells. There was a significant skewing with genes upregulated in Uchl1Tg GCBs disproportionately being enriched in light zone cells, whereas those downregulated are enriched in dark zone cells (Figure 7E). To further explore the impact of UCH-L1 on cell death, we used the TUNEL assay to examine apoptosis. Although wild-type and IμHABCL6 mice had similar numbers of TUNEL-positive cells in splenic follicles, both Uchl1 and Uchl1/IμHABCL6 double mutant mice had a significant decrease consistent with reduced cell death (Figure 7F). As apoptosis is a prominent event in the context of the GC reaction, we measured the output of the GC by measuring antigen-specific antibody responses following immunization with the T-cell–dependent antigen NP-KLH. In wild-type and Uchl1Tg mice, we find a steady increase in anti-NP IgG1 over 14 days (Figure 7G). In contrast, we find a significant reduction in the production of anti-NP IgG1 in IμHABCL6 mice. This impaired production of IgG1 is rescued by the Uchl1 transgene. Despite the defect in NP-specific IgG1 production, there is no significant difference in affinity maturation among the different genotypes.
Discussion
Here, we find that UCH-L1, an enzyme that was previously thought to be a specific marker of the neuroendocrine system, is induced in B cells undergoing the GC reaction and is also highly expressed in lymphomas derived from GCBs. Our prior work has revealed a novel role for UCH-L1 in regulating the prosurvival mTOR-AKT pathway both in malignant B cells and in the brain, where levels of UCH-L1 are highest. The ability of enforced UCH-L1 expression to promote spontaneous B-cell lymphoma, as well as those driven by MYC or BCL6, indicates that the prosurvival role of endogenous UCH-L1 in GCBs is rate-limiting, with higher levels able to further enhance B-cell survival and ultimately result in malignant transformation. Interestingly, our results with RNA interference suggest that in those GCB-DLBCL cells where UCH-L1 is high, it is providing an essential survival signal. The class of deubiquitinating enzymes is an emerging target for pharmaceutical compounds, with several inhibitors in preclinical development stages.26,27 An improved understanding of the role of UCH-L1 in disease and physiology provides important insight into the potential efficacy of small-molecule inhibitors for the treatment of B-cell cancers, and helps to understand the potential physiological effects of this strategy.
GCB-DLBCL disproportionately affects children, adolescents, and young adults, with the incidence of ABC subtype increasing with patient age.28-31 In adults, the outcome for patients with GCB disease is superior to that seen in patients with the ABC subtype.1 In patients of all ages, relapsed disease offers a much worse prognosis, with second-line therapies able to salvage only a fraction of patients.2,3,32-34 There are few clinical features or biomarkers that offer a robust risk-stratification option in DLBCL, particularly for those with GCB disease. Our data provide important insight into the ability to identify those with GCB disease at high risk for relapse. Future work is required to test the ability to detect high UCH-L1 on clinical samples using standard methods, and for this to predict relapse in a prospective manner.
Although this report focuses on the oncogenic role of UCH-L1 in the GC, our findings of strong induction of UCH-L1 in GCBs leads to questions about the transcriptional regulation and the potential physiological role of UCH-L1 in this compartment. Very little is known regarding the regulation of UCHL1, though 1 study has linked the PU.1 transcription factor (SPI1) with UCHL1 transcription in B-cell lymphoma.35 Unfortunately, the levels of UCHL1 do not correlate with those of SPI1 in the gene expression data sets we examined (data not shown). The level of UCHL1 transcript and protein in B cells transitioning through the GC mirror that of the master regulator BCL6, though there is no evidence that UCH-L1 levels are directly regulated by this factor. In fact, neither BCL6 depletion, nor the overexpression of degradation-deficient BCL6, substantially changed UCHL1 transcript levels.21,22 ChIP-seq data further do not demonstrate BCL6 occupancy of the UCHL1 promoter.23 It is of course possible that BCL6 acts indirectly to stimulate UCHL1 transcription. It will be important to further define the transcriptional regulation of UCHL1 in GCBs to better understand its role in the lymphomas arising from this structure.
Despite it being among the first deubiquitinating enzymes discovered,20 there is a paucity of data regarding its physiological roles. We recently reported a novel function of UCH-L1 in regulating mTOR-AKT signaling.8 High levels of enzyme drive increased assembly of mTORC2 that phosphorylates AKTS473, a function that requires catalytic activity. UCH-L1 antagonizes the activity of a CUL4-DDB1 ubiquitin E3 ligase in ubiquitinating the mTORC1 subunit raptor, with reduced ubiquitination seen in the presence of UCH-L1 or after the depletion of DDB1. Several prior studies have shown that AKT is hyperphosphorylated in a substantial proportion of DLBCL and Burkitt lymphoma.36-38 Although we have identified UCH-L1 to be one mechanism by which this may occur, there are, however, a number of other mechanisms leading to increased AKT activity in B-cell lymphoma. Recent sequencing studies have identified mutations in CD79A/B, GNA13, and S1PR2 genes that result in increased phosphatidylinositol 3-kinase (PI3K) activity in DLBCL,39,40 as well as loss of phosphatase and tensin homolog that interferes with its ability to restrain the PI3K-dependent generation of phosphatidylinositol 3,4,5 trisphosphate.36 Frequent mutations involving the ID3 and TCF3 genes also lead to increased PI3K activity in Burkitt lymphoma.41 The varied mechanisms that lead to AKT activation make it unlikely that UCH-L1 alone acts as a marker of higher sensitivity to AKT inhibitors, but our data further underscore the importance of this pathway in B-cell transformation. Our data regarding the ability of transgenic UCH-L1 to promote the production of antigen-specific class-switched antibody in mice with deregulated BCL6 expression suggest that it may also have a role in the humoral immune response. Further work is required to better understand the nature of this effect.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Ricardo Dalla-Favera (Columbia University, New York, NY) for providing IμHABCL6 mice and Dr P. Leif Bergsagel (Mayo Clinic Cancer Center, Scottsdale, AZ) for the Vk*MYC mice. The authors thank Drs Richard Bram, Anne Novak, and Rodney Miles for critical reading of the manuscript and Dr Farhad Kosari for assistance with gene expression data.
This work was supported by the National Institutes of Health National Cancer Institute (CA151351) (P.J.G.), the Multiple Myeloma Research Foundation (P.J.G.), Gabrielle’s Angel Foundation for Cancer Research (P.J.G.), the Hyundai Hope on Wheels Foundation (P.J.G.), and supported in part by the University of Iowa/Mayo Clinic Lymphoma SPORE CA97274. New gene expression data may be found through the Gene Expression Omninbus (http://www.ncbi.nlm.nih.gov/geo/) GSE76706.
P.J.G. is a former American Society of Hematology Basic Research Scholar, a past recipient of the Howard Hughes Medical Institute Physician Scientist Early Career Award, and a former Harriet H. Samuelsson Foundation Pediatric Cancer Research Scientist.
Authorship
Contribution: T.B. and S.H. performed the experiments; T.B., S.H., A.L.F., and P.J.G. interpreted experimental data; and P.J.G designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: P.J.G. receives research funding from Mission Therapeutics (Cambridge, United Kingdom) to conduct experiments that are not included in this manuscript. The remaining authors declare no competing financial interests.
Correspondence: Paul J. Galardy, Mayo Clinic, Guggenheim 15, 200 First St SW, Rochester, MN 55905; e-mail: galardy.paul@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal