Key Points
CDKN2A and TP53 deletions remain of bad prognostic value in younger MCL patients treated according to the current standard of care.
CDKN2A and TP53 deletions have independent deleterious effects and should be considered for treatment decisions in addition to MIPI and Ki-67 index.
Abstract
We revisited the prognostic value of frequently detected somatic gene copy number alterations (CNAs) in mantle cell lymphoma (MCL) patients treated first line with immunochemotherapy and autologous stem cell transplantation (ASCT), with or without high-dose cytarabine, in the randomized European MCL Younger trial. DNA extracted from tumor material of 135 patients (median age, 56 years) was analyzed by multiplex ligation-dependent probe amplification and/or quantitative multiplex polymerase chain reaction of short fluorescent fragments. As expected, MYC (18%) was the more frequently gained, whereas RB1 (26%), ATM (25%), CDKN2A (p16) (25%), and TP53 (22%) were the more frequently deleted. Whether adjusted for MCL International Prognostic Index (MIPI) or not, deletions of RB1, CDKN2A, TP53, and CDKN1B were associated with shorter overall survival (OS), similarly in both treatment arms, whereas CNAs in MYC, ATM, CDK2, CDK4, and MDM2 had no prognostic value. Additive effects were seen for CDKN2A (hazard ratio, 2.3; P = .007, MIPI-adjusted) and TP53 deletions (hazard ratio, 2.4; P = .007), reflected in a dismal outcome with simultaneous deletions (median OS, 1.8 years) compared with single deletions (median OS, 4.3 and 5.1 years) or without these deletions (median OS, 7 years), again similarly in both treatment arms. The additive prognostic effects of CDKN2A and TP53 deletions were independent of the Ki-67 index. Despite immunochemotherapy, high-dose cytarabine, and ASCT, younger MCL patients with deletions of CDKN2A (p16) and TP53 show an unfavorable prognosis and are candidates for alternative therapeutic strategies. This trial was registered at www.clinicaltrials.gov as #NCT00209222.
Introduction
Mantle cell lymphoma (MCL) is a well-defined subtype of B-cell lymphoma characterized by t(11;14)(q13;q32)-driven overexpression of cyclin D1.1,2 It is rare, representing only 5% to 9% of all lymphomas.3 The median age at diagnosis is ∼65 years. Other than occasional indolent forms,4 MCL is considered to be an aggressive disease, and its treatment is a challenge.5,6 Despite considerable improvement in progression-free survival7 (∼2 years after first-line conventional immunochemotherapy and >5 years after dose-intensified therapy including high-dose cytarabine8 ), individual heterogeneity in clinical behavior is still encountered, ranging from primary refractory disease to a progression-free survival of >7 years.9
Although numerous genetic abnormalities are involved in the pathophysiology of MCL,10,11 not all are of prognostic value. The CDKN2A locus (9p21), which encodes for both the CDK4/6 inhibitor INK4a (p16) and the positive TP53 regulator ARF (p14), is one of the most frequently deleted and is consistently associated with a poor prognosis in MCL.12-19 Additional genes frequently targeted by secondary genetic alterations,20 eg, CDKN1B/CDK2/CDK4/RB1, involved in cell cycle regulation,16,17,21,22 and ATM/MDM2/TP53, involved in DNA damage response,14,18,21 are less consistently associated with prognosis. Alterations of the 8q24 (MYC) locus are uncommon in MCL but have been identified in cases with blastoid MCL.23,24 The association of the overexpression of the primary cell cycle regulator cyclin D1 and secondary alterations in cell cycle regulation translates into high level cell proliferation and shorter survival.13 Accordingly, the tumor cell proliferation marker Ki-67 index25,26 adds to the prognostic value of the MCL International Prognostic Index (MIPI),27 the first clinical prognostic index specific for MCL.28
Previous reports on the prognostic value of gene copy number alterations (CNAs) in MCL are mainly based on smaller retrospective patient cohorts and/or with heterogeneous treatment, including only 0% to 40% of patients with rituximab-containing chemotherapy. Using the clinical data of the randomized European MCL Younger trial,29 the aim of our study was to re-evaluate the prognostic value of somatic CNAs in the context of the current standard of care, including immunochemotherapy, high-dose cytarabine, and autologous stem cell transplantation (ASCT).8
Patients and methods
Patients
Patients up to 65 years old with previously untreated MCL of Ann Arbor stages II to IV were enrolled in the randomized MCL Younger trial of the European MCL Network (#NCT00209222). Patients received either 6 cycles of rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), followed by stem cell mobilization and myeloablative radio-chemotherapy with ASCT, or 6 cycles of alternating R-CHOP or rituximab, dexamethasone, high-dose cytarabine, and cisplatin (R-DHAP), followed by high-dose cytarabine containing myeloablative radio-chemotherapy and ASCT29 (supplemental Figure 1 available on the Blood Web site). The trial was performed in accordance with the Declaration of Helsinki, and all patients gave written informed consent to participate and to provide blood, bone marrow, and tissue for biologic studies.
The inclusion criteria for patients analyzed in the present biological study were (1) a confirmed histologic diagnosis of MCL with overexpression of cyclin D1 assessed by immunohistochemistry and/or molecular biology; (2) the availability of diagnostic tumor DNA; and (3) complete clinical data. When no frozen tissue DNA was available, peripheral samples with >50% tumor cells were considered eligible for CNA analysis. Diagnostic peripheral blood and bone marrow aspirations had been prospectively collected during the minimal residual disease program linked to the MCL Younger trial,30 with tumor load prospectively assessed by 4-color flow cytometry and/or clonospecific quantitative polymerase chain reaction (PCR). Finally, when no other diagnostic material was available, formalin-fixed, paraffin-embedded (FFPE) tumor sections were collected from the LYSA-Pathology platform.
Histopathologic and clinical variables
The histologic diagnosis was confirmed by the central pathology review at one of the designated pathology reference centers (European MCL Pathology Panel). MCL cytology was classified as blastoid (pleomorphic or blastic) or nonblastoid (classical or small cell).31 The Ki-67 index was defined as the percentage of Ki-67–positive tumor cells in representative areas of the lymphoma and quantified according to published guidelines.32 We used the established 30% threshold to distinguish patients with a low or high Ki-67 index.33 The individual clinical prognostic information was summarized in the quantitative MIPI score using age, performance status, lactate dehydrogenase (LDH), and white blood cell count (WBC) at trial entry.27 Outcome variables were overall survival (OS) and time to treatment failure (TTF). OS was measured from trial registration to death from any cause. Patients who had not died during the observation period were censored for OS at the latest contact date. TTF was measured from start of treatment to stable disease at end of induction, progression, or death from any cause. Patients with no failure, progression, or death during observation time were censored for TTF at the latest tumor assessment date. The clinical data cutoff date was April 7, 2013.
DNA extraction and quality control
DNA was extracted from peripheral blood and/or bone marrow samples using standard proteinase K digestion and phenol-chloroform extraction or the QIAGEN Blood Mini Kit (QIAGEN).30 DNAs extracted from tumor tissues were sent to the Creteil biological center by French locoregional “Centres de Ressources Biologiques.” The quality of tumor DNA samples was assessed using the BIOMED-2 specimen control “barcode“ multiplex PCR reaction.34
CNA analysis
Quantitative multiplex PCR of short fluorescent fragments
An MCL-dedicated quantitative multiplex PCR of short fluorescent fragments (QMPSF) assay was used as described (supplemental Figure 2A).18 The following target genes were analyzed: MYC (8q24), RB1 (13q14), ATM (11q22-23), CDKN2A (p16) (9p21), TP53 (17p13), CDK2 (12q13), CDKN1B (p27) (12p13), and MDM2 (12q14). The reference gene was CECR1, located at 22q11. Two control DNAs were used to calculate the mean template/control peak height ratio (commercial DNA; Roche Diagnostics, Mylan, France). PCR fragments ranged from 150 to 250 bp. QMPSF is able to detect CNAs when tumor cells represent >30% of total cells analyzed.18
Multiplex ligation-dependent probe amplification
In contrast to QMPSF, multiplex ligation-dependent probe amplification (MLPA) (supplemental Figure 2A) probe-targeted sequences are small (50-70 nucleotides), an attractive property for FFPE-derived analysis. Hybridization and ligation steps were performed on 100 ng genomic DNA, and probes were subjected to PCR reactions using SALSA P038-A2 CLL probemix-2 according to the manufacturer's instructions (MRC-Holland, Amsterdam, The Netherlands). Targeted loci included 8q24 (MYC), several targets on 13q14, including one specific for RB1, 9p21 (CDKN2A [p16]), 11q23 (ATM), 17p13 (TP53), and 12q14 (CDK4). Products were run on an automated sequencer (ABI 3130X1 genetic analyzer). The calculation of probe ratios consisted of a mathematical comparison between relative quantities of target DNA amplified from a test patient sample and those generated in 6 normal control samples. It was performed using Coffalyser software (MRC-Holland BV) A decrease or increase in peak height values resulting in ratios <0.80 or >1.20, respectively, indicated copy loss or gain in tumor cells. MLPA has been reported to detect CNAs when tumor cells represent >40% of total cells analyzed.35,36 In a validation step using the GRANTA cell line, we were able to detect the homozygous CDKN2A (p16) deletion when only 25% of tumor cells were present in studied samples (sample/reference allele ratio = 0.5; supplemental Figure 2B).
CNA result encoding
The first technique used was QMPSF. However, this technique was not suited for paraffin-embedded tissues, and several patients had an alteration of the gene used for normalization. We moved to the MLPA technique and reanalyzed all patients for whom enough tumor material remained available. From 560 comparisons for shared loci, 13 discrepancies between the 2 techniques were observed (supplemental Figure 2C). In discrepant cases, MLPA was repeated. If another repeat could not be performed (no more material), results were considered inconclusive. When repeated, MLPA confirmed precedent MLPA results in all performed cases, and the obtained MLPA values were considered for analyses. For samples with an estimated 50% tumor cell infiltration and a sample/reference ratio between 0.8 and 0.9, results were reported as inconclusive to avoid false negativities.
Statistical methods
Clinical characteristics, histopathologic features, and clinical outcome of CNA evaluable MCL trial patients were compared with nonevaluable patients. Univariable analyses of the prognostic value of CNAs were done using Kaplan-Meier estimates and log-rank tests. For all genes with CNAs detected in >10 patients, we performed univariable and multivariable Cox regression analyses. We adjusted for clinical prognostic factors summarized in the quantitative MIPI score and for the Ki-67 index. In multivariable analyses, we identified independently prognostic CNAs by backward elimination using the Wald statistic, keeping MIPI score constantly included. Initially, all CNAs with prognostic impact in univariable analyses, indicated by P < .05, were included in multiple Cox regression models, and subsequently backward elimination was performed using an exclusion level of P < .01. As sensitivity analyses, we added all CNAs not included in multivariable analyses for backward elimination separately to the final Cox model.
We performed subgroup analyses according to treatment group and investigated the potential interaction of prognostic effects with treatment using interaction terms in multivariable Cox regression. For statistical comparisons, we used Fisher’s exact, Mann-Whitney U, log-rank, and Wald tests. All reported P values are 2 sided and descriptive.
Results
Patient characteristics
One hundred thirty-five patients from the MCL Younger trial had suitable tumor DNA available for the CNA study (supplemental Figure 3); 72 patients were in the experimental R-CHOP/R-DHAP group. The median age at trial registration was 56 years (Table 1). Eighty-six percent of patients had stage IV disease, and according to MIPI, 49%, 26%, and 25% of patients were of low, intermediate, and high risk, respectively. Eight (9%) of 89 patients with assessed cytology had blastoid MCL. The median Ki-67 index was 19%, and the Ki-67 index was ≥30% in 32% of the 72 patients with an available Ki-67 index. After a median follow-up of 4.6 years, the estimated median OS and TTF for the 135 patients was 6.8 (50 events) and 4.6 years (72 events), respectively. Compared with the 440 MCL patients not included in the biologic study, selected patients had similar age, more frequent bone marrow involvement, increased LDH, and higher WBC and thus had a less favorable risk profile (Table 1). Accordingly, OS and TTF were slightly shorter in the studied patients compared with the nonstudied patients (5-year OS rates, 66% vs 73%; P = .0083; 5-year TTF rates, 46% vs 55%; P = .0122).
Patient baseline characteristics
| Variable . | Value . | Included (n = 135) . | Not included (n = 440) . | P value . |
|---|---|---|---|---|
| Age (years) | Median (min-max) | 56 (35-65) | 55 (30-66) | .33 |
| Male | No. (%) | 109 (81) | 350 (80) | .81 |
| Stage | .55 | |||
| I | No. (%) | 0 (0) | 2 (0) | |
| II | No. (%) | 3 (2) | 20 (5) | |
| III | No. (%) | 16 (12) | 61 (14) | |
| IV | No. (%) | 116 (86) | 357 (81) | |
| B-symptoms | No. (%) | 51 (38) | 163 (37) | .92 |
| ECOG PS | .061 | |||
| 0-1 | No. (%) | 126 (93) | 428 (97) | |
| 2 | No. (%) | 9 (7) | 12 (3) | |
| Bone marrow involvement | No. (%) | 113 (84) | 323 (73) | .016 |
| Elevated LDH | No. (%) | 63 (47) | 147 (33) | .006 |
| LDH/ULN | Median (min-max) | 0.97 (0.41-4.8) | 0.89 (0.29-12.2) | .038 |
| WBC (G/L) | Median (min-max) | 8.8 (0.052-388) | 7.2 (1.1-1110) | .001 |
| MIPI score | Median (min-max) | 5.73 (3.63-8.03) | 5.51 (4.07-8.68) | <.001 |
| MIPI risk | <.001 | |||
| Low | No. (%) | 66 (49) | 294 (67) | |
| Intermediate | No. (%) | 35 (26) | 96 (22) | |
| High | No. (%) | 34 (25) | 50 (11) | |
| Ki-67 index (%)* | Median (min-max) | 19 (2-87) | 21 (0-97) | .52 |
| Ki-67 index ≥ 30% | No. (%) | 23 (32) | 63 (26) | .30 |
| Blastoid cytology† | No. (%) | 8 (9) | 22 (8) | .32 |
| Treatment | .83 | |||
| R-CHOP | No. (%) | 63 (47) | 174 (44)‡ | |
| R-CHOP/R-DHAP | No. (%) | 72 (53) | 217 (55)‡ |
| Variable . | Value . | Included (n = 135) . | Not included (n = 440) . | P value . |
|---|---|---|---|---|
| Age (years) | Median (min-max) | 56 (35-65) | 55 (30-66) | .33 |
| Male | No. (%) | 109 (81) | 350 (80) | .81 |
| Stage | .55 | |||
| I | No. (%) | 0 (0) | 2 (0) | |
| II | No. (%) | 3 (2) | 20 (5) | |
| III | No. (%) | 16 (12) | 61 (14) | |
| IV | No. (%) | 116 (86) | 357 (81) | |
| B-symptoms | No. (%) | 51 (38) | 163 (37) | .92 |
| ECOG PS | .061 | |||
| 0-1 | No. (%) | 126 (93) | 428 (97) | |
| 2 | No. (%) | 9 (7) | 12 (3) | |
| Bone marrow involvement | No. (%) | 113 (84) | 323 (73) | .016 |
| Elevated LDH | No. (%) | 63 (47) | 147 (33) | .006 |
| LDH/ULN | Median (min-max) | 0.97 (0.41-4.8) | 0.89 (0.29-12.2) | .038 |
| WBC (G/L) | Median (min-max) | 8.8 (0.052-388) | 7.2 (1.1-1110) | .001 |
| MIPI score | Median (min-max) | 5.73 (3.63-8.03) | 5.51 (4.07-8.68) | <.001 |
| MIPI risk | <.001 | |||
| Low | No. (%) | 66 (49) | 294 (67) | |
| Intermediate | No. (%) | 35 (26) | 96 (22) | |
| High | No. (%) | 34 (25) | 50 (11) | |
| Ki-67 index (%)* | Median (min-max) | 19 (2-87) | 21 (0-97) | .52 |
| Ki-67 index ≥ 30% | No. (%) | 23 (32) | 63 (26) | .30 |
| Blastoid cytology† | No. (%) | 8 (9) | 22 (8) | .32 |
| Treatment | .83 | |||
| R-CHOP | No. (%) | 63 (47) | 174 (44)‡ | |
| R-CHOP/R-DHAP | No. (%) | 72 (53) | 217 (55)‡ |
ECOG PS, Eastern Cooperative Oncology Group performance status; ULN, upper limit of normal range.
Ki-67 assessed in 72 of 135 included and in 245 of 440 not included patients.
Cytology assessed in 68 of 135 included and 270 of 440 not included patients.
In 48 patients treatment not documented, 1 R-bendamustin.
CNA detection
DNA with >50% tumor cells was obtained from lymph node biopsies for 79 patients (including 61 frozen and 18 FFPE samples), peripheral blood for 54 patients, and bone marrow aspirates for 2 patients. The most frequent copy gain involved MYC (18%; Table 2), whereas the most frequent deletion involved the 13q14 locus (36%), including RB1 in 26%. CNAs in ATM were mostly deletions (25%), but copy gains were found in 3 of 129 patients. As expected, CDKN2A (p16) and TP53 deletions were frequent (25% and 22%, respectively). Of 114 patients analyzed with both QMPSF and MLPA, only 24 (21%) displayed no CNA in any analyzed loci. Details of the CNA distribution for each MCL case are shown in supplemental Figure 4. The distribution of CNAs did not differ substantially according to the type of sample analyzed (supplemental Table 1).
Distribution of gene copy number alterations in the studied cohort
| Allelic status . | MYC (8q24) . | Locus (13q14) . | RB1 (13q14) . | ATM (11q23) . | CDKN2A (9p21) . | TP53 (17p13) . | CDK2 (12q13) . | CDK4 (12q14) . | CDKN1B (12p13) . | MDM2 (12q14) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Total interpretable (n) | 130 | 130 | 131 | 129 | 134 | 134 | 116 | 129 | 109 | 114 |
| Diploid locus (n) | 107 | 82 | 96 | 94 | 100 | 104 | 106 | 119 | 93 | 108 |
| Diploid locus (%) | 82% | 63% | 73% | 73% | 75% | 78% | 91% | 92% | 85% | 95% |
| Monoallelic deletion (n) | 0 | 44 | 33 | 31 | 19 | 30 | 1 | 2 | 13 | 2 |
| Monoallelic deletion (%) | 0% | 34% | 25% | 24% | 14% | 22% | 1% | 2% | 12% | 2% |
| Biallelic deletion (n) | 0 | 3 | 1 | 1 | 15 | 0 | 0 | 0 | 0 | 0 |
| Biallelic deletion (%) | 0% | 2% | 1% | 1% | 11% | 0% | 0% | 0% | 0% | 0% |
| Gain (n) | 23 | 1 | 1 | 3 | 0 | 0 | 9 | 8 | 3 | 4 |
| Gain (%) | 18% | 1% | 1% | 2% | 0% | 0% | 8% | 6% | 3% | 4% |
| Total deletions (%) | 0% | 36% | 26% | 25% | 25% | 22% | 1% | 2% | 12% | 2% |
| Total abnormalities (%) | 18% | 37% | 27% | 27% | 25% | 22% | 9% | 8% | 15% | 5% |
| Allelic status . | MYC (8q24) . | Locus (13q14) . | RB1 (13q14) . | ATM (11q23) . | CDKN2A (9p21) . | TP53 (17p13) . | CDK2 (12q13) . | CDK4 (12q14) . | CDKN1B (12p13) . | MDM2 (12q14) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Total interpretable (n) | 130 | 130 | 131 | 129 | 134 | 134 | 116 | 129 | 109 | 114 |
| Diploid locus (n) | 107 | 82 | 96 | 94 | 100 | 104 | 106 | 119 | 93 | 108 |
| Diploid locus (%) | 82% | 63% | 73% | 73% | 75% | 78% | 91% | 92% | 85% | 95% |
| Monoallelic deletion (n) | 0 | 44 | 33 | 31 | 19 | 30 | 1 | 2 | 13 | 2 |
| Monoallelic deletion (%) | 0% | 34% | 25% | 24% | 14% | 22% | 1% | 2% | 12% | 2% |
| Biallelic deletion (n) | 0 | 3 | 1 | 1 | 15 | 0 | 0 | 0 | 0 | 0 |
| Biallelic deletion (%) | 0% | 2% | 1% | 1% | 11% | 0% | 0% | 0% | 0% | 0% |
| Gain (n) | 23 | 1 | 1 | 3 | 0 | 0 | 9 | 8 | 3 | 4 |
| Gain (%) | 18% | 1% | 1% | 2% | 0% | 0% | 8% | 6% | 3% | 4% |
| Total deletions (%) | 0% | 36% | 26% | 25% | 25% | 22% | 1% | 2% | 12% | 2% |
| Total abnormalities (%) | 18% | 37% | 27% | 27% | 25% | 22% | 9% | 8% | 15% | 5% |
CNAs, Ki-67 index, and cytology
Among the patients with high Ki-67 index (≥30%), deletions in 13q14, RB1, CDKN2A (p16), and CDKN1B (p27) were substantially more frequent compared with patients with low Ki-67 index (65% vs 25%, 48% vs 19%, 43% vs 10%, and 24% vs 8%, respectively), whereas the frequencies of the remaining CNAs, including TP53 deletion, were independent of the Ki-67 index (22% vs 18%; supplemental Table 2). Frequencies of CNAs were similar in blastoid vs nonblastoid MCL, except for TP53, which was more frequently deleted in blastoid MCL (4 of 8) compared with nonblastoid MCL (11 of 81, 14%; supplemental Table 2).
Prognostic relevance of CNAs
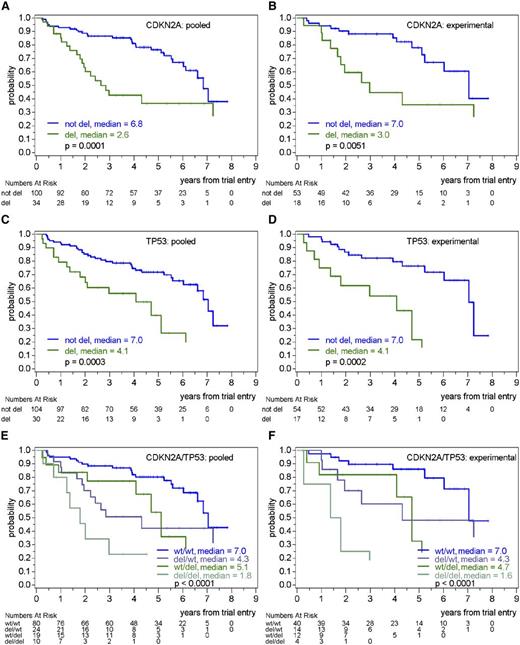
In analyses including single genes without and with adjustment for MIPI, deletions of the 13q14 locus, RB1, CDKN2A (p16), TP53, and CDKN1B (p27) were associated with shorter OS and TTF (Table 3; Figure 1; supplemental Figure 5), whereas CNAs in MYC, ATM, CDK2, CDK4, and MDM2 were not prognostic (Table 3). The MIPI-adjusted OS hazard ratio (HR) for deletions in the 13q14 locus was 2.1 (P = .010), whereas it was 2.2 (P = .014) for RB1 deletions and 2.2 (P = .010) for CDKN2A (p16) deletions. Patients with heterozygous or homozygous deletions of CDKN2A (p16) showed a similar outcome. For TP53 deletions, the adjusted OS HR was 2.4 (P = .008) and for CDKN1B (p27) deletions was 1.9 (P = .083). Similar results were seen for TTF (Table 3). There was no indication for an interaction of the prognostic effect for any CNA with the treatment group. In particular, deletions of CDKN2A (p16) or TP53 had similar prognostic value in the experimental (Figure 1) and control treatment groups (supplemental Figures 5 and 6).
Prognostic impact of CNAs by Cox regression analyses without or with adjustment for MIPI score
| Variable . | Adjustment . | OS . | TTF . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI, lower bound . | 95% CI, upper bound . | P . | HR . | 95% CI, lower bound . | 95% CI, upper bound . | P . | ||
| MYC gain | None | 0.89 | 0.43 | 1.84 | .74 | 1.01 | 0.56 | 1.82 | .98 |
| MIPI score | 1.03 | 0.49 | 2.16 | .93 | 1.10 | 0.61 | 2.00 | .75 | |
| 13q14 deletion | None | 2.03 | 1.15 | 3.60 | .015 | 2.40 | 1.47 | 3.92 | <.001 |
| MIPI score | 2.13 | 1.19 | 3.80 | .010 | 2.49 | 1.52 | 4.06 | <.001 | |
| RB1 deletion | None | 1.75 | 0.96 | 3.22 | .070 | 1.91 | 1.14 | 3.19 | .013 |
| MIPI score | 2.20 | 1.18 | 4.11 | .014 | 2.25 | 1.33 | 3.79 | .002 | |
| ATM deletion | None | 0.65 | 0.33 | 1.29 | .22 | 0.81 | 0.47 | 1.38 | .44 |
| MIPI score | 0.77 | 0.38 | 1.55 | .47 | 0.91 | 0.52 | 1.57 | .72 | |
| CDKN2A deletion | None | 2.97 | 1.67 | 5.29 | <.001 | 2.62 | 1.59 | 4.33 | <.001 |
| MIPI score | 2.22 | 1.21 | 4.07 | .010 | 2.15 | 1.28 | 3.62 | .004 | |
| TP53 deletion | None | 2.95 | 1.58 | 5.48 | .001 | 2.39 | 1.41 | 4.06 | .001 |
| MIPI score | 2.36 | 1.26 | 4.42 | .008 | 1.96 | 1.14 | 3.36 | .015 | |
| CDKN1B deletion | None | 2.31 | 1.13 | 4.70 | .021 | 2.42 | 1.28 | 4.57 | .007 |
| MIPI score | 1.89 | 0.92 | 3.89 | .083 | 2.03 | 1.07 | 3.85 | .031 | |
| Variable . | Adjustment . | OS . | TTF . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI, lower bound . | 95% CI, upper bound . | P . | HR . | 95% CI, lower bound . | 95% CI, upper bound . | P . | ||
| MYC gain | None | 0.89 | 0.43 | 1.84 | .74 | 1.01 | 0.56 | 1.82 | .98 |
| MIPI score | 1.03 | 0.49 | 2.16 | .93 | 1.10 | 0.61 | 2.00 | .75 | |
| 13q14 deletion | None | 2.03 | 1.15 | 3.60 | .015 | 2.40 | 1.47 | 3.92 | <.001 |
| MIPI score | 2.13 | 1.19 | 3.80 | .010 | 2.49 | 1.52 | 4.06 | <.001 | |
| RB1 deletion | None | 1.75 | 0.96 | 3.22 | .070 | 1.91 | 1.14 | 3.19 | .013 |
| MIPI score | 2.20 | 1.18 | 4.11 | .014 | 2.25 | 1.33 | 3.79 | .002 | |
| ATM deletion | None | 0.65 | 0.33 | 1.29 | .22 | 0.81 | 0.47 | 1.38 | .44 |
| MIPI score | 0.77 | 0.38 | 1.55 | .47 | 0.91 | 0.52 | 1.57 | .72 | |
| CDKN2A deletion | None | 2.97 | 1.67 | 5.29 | <.001 | 2.62 | 1.59 | 4.33 | <.001 |
| MIPI score | 2.22 | 1.21 | 4.07 | .010 | 2.15 | 1.28 | 3.62 | .004 | |
| TP53 deletion | None | 2.95 | 1.58 | 5.48 | .001 | 2.39 | 1.41 | 4.06 | .001 |
| MIPI score | 2.36 | 1.26 | 4.42 | .008 | 1.96 | 1.14 | 3.36 | .015 | |
| CDKN1B deletion | None | 2.31 | 1.13 | 4.70 | .021 | 2.42 | 1.28 | 4.57 | .007 |
| MIPI score | 1.89 | 0.92 | 3.89 | .083 | 2.03 | 1.07 | 3.85 | .031 | |
By log-rank tests, no prognostic effects were seen for CNAs in CDK2 (P = .87 for TTF and P = .45 for OS), CDK4 (P = .34 for TTF and P = .14 for OS), and MDM2 (P = .43 for TTF and P = .69 for OS). Due to the low number of CNAs, these genes were not included in Cox regression analyses. CI, confidence interval.
Prognostic value of CDKN2A (p16) and TP53 deletions on OS in pooled treatment arms or in the experimental treatment arm (R-CHOP/R-DHAP). (A) OS according to the presence or absence of CDKN2A (p16) deletion in pooled treatment arms. (B) OS according to the presence or absence of CDKN2A (p16) deletion in the experimental treatment arm. (C) OS according to the presence or absence of TP53 deletion in pooled treatment arms. (D) OS according to the presence or absence of TP53 deletion in the experimental treatment arm. (E) OS according to the presence or absence of CDKN2A (p16) and TP53 deletions in pooled treatment arms. (F) OS according to the presence or absence of CDKN2A (p16) and TP53 deletions in the experimental treatment arm.
Prognostic value of CDKN2A (p16) and TP53 deletions on OS in pooled treatment arms or in the experimental treatment arm (R-CHOP/R-DHAP). (A) OS according to the presence or absence of CDKN2A (p16) deletion in pooled treatment arms. (B) OS according to the presence or absence of CDKN2A (p16) deletion in the experimental treatment arm. (C) OS according to the presence or absence of TP53 deletion in pooled treatment arms. (D) OS according to the presence or absence of TP53 deletion in the experimental treatment arm. (E) OS according to the presence or absence of CDKN2A (p16) and TP53 deletions in pooled treatment arms. (F) OS according to the presence or absence of CDKN2A (p16) and TP53 deletions in the experimental treatment arm.
In multivariable analyses including deletions of RB1, CDKN2A (p16), TP53, and CDKN1B (p27) and adjusting for MIPI, only deletions of CDKN2A (p16) (OS HR, 2.3; P = .007) and TP53 (OS HR, 2.4; P = .007) showed independent prognostic impact. The group with both CDKN2A (p16) and TP53 deletions showed the worst prognosis, with median OS and TTF of 1.8 and 0.5 years, respectively (Figure 1; supplemental Figure 5), and again, as there was no interaction with the treatment group, the independent prognostic impact of the CDKN2A (p16) deletion and TP53 deletion on OS and TTF was clearly seen in both the experimental (R-CHOP/R-DHAP) arm (Figure 1; supplemental Figure 5) and the control (R-CHOP) arm (supplemental Figure 6). No other CNA investigated showed an additional prognostic impact to CDKN2A (p16) and TP53 deletions.
When adjusted for the Ki-67 index, the prognostic impact of CDKN2A (p16) deletion as single variable on OS still persisted but was reduced (HR unadjusted, 5.3; adjusted for Ki-67 index, 3.8; adjusted for Ki-67 index and MIPI, 3.6), whereas the prognostic impact of TP53 deletion as single variable was only slightly modified by the inclusion of the Ki-67 index (HR unadjusted, 2.7; adjusted for Ki-67 index without or with MIPI, 2.4). In multivariable models including CDKN2A (p16), TP53, and the Ki-67 index on 72 patients with available Ki-67 index (26 events), without or with adjustment for MIPI, the independent prognostic effect on OS of deletions in CDKN2A (p16) (HR, 4.2 without or with adjustment for MIPI) and TP53 (HR, 2.5 and 2.8 without and with adjustment for MIPI) was shown to be independent of the Ki-67 index (Table 4).
Multivariable Cox regression analyses for overall survival including CDKN2A deletion and TP53 deletion status, excluding or including Ki-67 index (in %), without or with adjustment for MIPI score
| Variable . | Adjustment . | OS . | |||
|---|---|---|---|---|---|
| HR . | 95% CI (lower bound) . | 95% CI, upper bound . | P . | ||
| CDKN2A deletion | None | 5.51 | 2.42 | 12.54 | <.001 |
| TP53 deletion | 2.92 | 1.23 | 6.93 | .015 | |
| CDKN2A deletion | MIPI score | 4.46 | 1.85 | 10.78 | .001 |
| TP53 deletion | 2.86 | 1.20 | 6.80 | .017 | |
| CDKN2A deletion | None | 4.15 | 1.55 | 11.14 | .005 |
| TP53 deletion | 2.54 | 1.04 | 6.24 | .041 | |
| Ki-67 index (10% increase) | 1.11 | 0.92 | 1.33 | .27 | |
| CDKN2A deletion | MIPI score | 4.19 | 1.50 | 11.66 | .006 |
| TP53 deletion | 2.79 | 1.15 | 6.78 | .024 | |
| Ki-67 index (10% increase) | 1.03 | 0.83 | 1.28 | .81 | |
| Variable . | Adjustment . | OS . | |||
|---|---|---|---|---|---|
| HR . | 95% CI (lower bound) . | 95% CI, upper bound . | P . | ||
| CDKN2A deletion | None | 5.51 | 2.42 | 12.54 | <.001 |
| TP53 deletion | 2.92 | 1.23 | 6.93 | .015 | |
| CDKN2A deletion | MIPI score | 4.46 | 1.85 | 10.78 | .001 |
| TP53 deletion | 2.86 | 1.20 | 6.80 | .017 | |
| CDKN2A deletion | None | 4.15 | 1.55 | 11.14 | .005 |
| TP53 deletion | 2.54 | 1.04 | 6.24 | .041 | |
| Ki-67 index (10% increase) | 1.11 | 0.92 | 1.33 | .27 | |
| CDKN2A deletion | MIPI score | 4.19 | 1.50 | 11.66 | .006 |
| TP53 deletion | 2.79 | 1.15 | 6.78 | .024 | |
| Ki-67 index (10% increase) | 1.03 | 0.83 | 1.28 | .81 | |
Discussion
We report here the prognostic value of CNAs detected at diagnosis in MCL patients treated in a randomized trial according to current standard of care, including a high-dose cytarabine regimen in the experimental arm.9 We show that deletions of CDKN2A (p16) and TP53 are of poor prognostic value for both OS and TTF, independently of MIPI and that CDKN2A (p16) and TP53 deletions are independent additive poor prognostic factors in both treatment arms. Moreover, we observed that the prognostic effects of CDKN2A (p16) and TP53 deletions were independent of the proliferation marker Ki-67 index.
Our CNA frequencies are in line with previously reported abnormalities detected by accurate techniques such as array or matrix comparative genomic hybridization (CGH), fluorescence in situ hybridization, real-time quantitative polymerase chain reaction (RQ-PCR), or QMPSF (supplemental Table 3). Deletions in 13q14 and ATM (11q23) are consistently the most frequently detected and in the same order of magnitude, followed by CDKN2A (9p21) and TP53 deletions.
We found that 22% of the studied patients had a TP53 deletion. The prognostic value of this abnormality is controversial in the MCL literature, with reports either showing no or a deleterious prognostic value (supplemental Table 3). However, these mostly smaller studies mixed patients with no need for treatment and patients with very aggressive disease. Moreover, treatment was heterogeneous, with <50% of patients receiving rituximab in combination with chemotherapy at induction. To our knowledge, we show here for the first time that the addition of rituximab to chemotherapy does not erase the poor prognostic value of TP53 deletion in MCL, despite a clinical improvement of disease outcome.37 The same has recently been observed in B-cell chronic lymphocytic leukemia.38 Our data from a randomized trial enable us to further demonstrate that induction treatment including high-dose cytarabine followed by ASCT, the new standard of care for younger patients, does not solve the problem of TP53 deletions for MCL patients. This is in accordance with Nordström et al, who recently demonstrated a poor prognostic value of p53 overexpression, as assessed by immunohistochemistry, in MCL patients treated with intensive immunochemotherapy including high-dose cytarabine.39
Most of our studied genes can be grouped into 2 major pathways, related to either cell cycle regulation (CDKN1B [p27], CDK4, and RB1) or apoptosis and DNA damage response pathways (ATM/TP53/MDM2). The CDKN2A (p16) deletion is thought to alter both cell cycle regulation (p16) and DNA damage response (p14) pathways in MCL. Previous reports in aggressive lymphomas of various histology40,41 or focusing on DLBCL42 have shown that reinforcing the p14-induced deficit of apoptosis and DNA damage response pathways through TP53 deletion, in addition to CDKN2A (p16) deletion, has an additional deleterious effect on OS. We now confirm these observations in the era of new treatments and in a larger cohort of MCL patients, highlighting the biologic power of these oncogenic alterations that are not overrun yet by the new therapeutic strategies.
Those 2 major alterations that accelerate cell cycle and inhibit apoptosis can be targeted by new therapies. Recent phase 2 studies have shown high response rates of small molecules targeting the B-cell receptor pathway, including Bruton tyrosine kinase inhibitors (such as ibrutinib)43 and phosphatidylinositol-4,5-bisphosphate 3-kinase inhibitors (like idelalisib),44 as well as proteasome inhibitors (bortezomib)45 and the immunomodulatory agents (lenalidomide).46 Based on the involved pathways, it would be logical to evaluate combined approaches targeting the different pathways. In fact, preclinical data suggest that the combination of ibrutinib and ABT-19947 or ibrutinib and bortezomib48,49 has synergistic efficacy. In addition, refractoriness to one or another compound may be overcome by such a combined approach.48
The population we studied for CNAs did not fully reflect the total population of the clinical trial, as bone marrow involvement, elevated LDH, and high-risk MIPI were more frequently observed. Because our patients were selected by the availability of a high content tumor cell sample, 42% had >50% circulating tumor cells. This selection could have affected the frequency of CNAs reported here. However, apart from the CDK2 and CDK4 gains, which tended to be more frequently detected in lymph nodes than in peripheral blood, no difference was detected between the 2 sample types analyzed. In particular, the deletion of CDKN2A (p16), which is associated with a proliferative signature,13 is not higher in frankly leukemic forms. Accordingly, the Ki-67 index was similar in studied and unstudied patients. We did not detect an additive prognostic impact of any other CNA investigated to CDKN2A (p16) and TP53 deletions. However, although our cohort of homogenously treated MCL patients analyzed for CNAs is relatively large compared with other reports, an even larger statistical power might be necessary to detect a prognostic impact especially of the more uncommon CNAs.
In conclusion, despite being associated with substantial improvement in clinical outcome, high-dose cytarabine as part of immunochemotherapy followed by ASCT does not seem to erase the bad prognostic value of CDKN2A (p16) and TP53 deletions. Furthermore, patients with simultaneous deletions of CDKN2A (p16) and TP53 show a very dismal outcome and are therefore candidates for specific therapeutic strategies. We therefore recommend that all patients benefit from CNA evaluation of CDKN2A and TP53 at diagnosis, in addition to Ki-67 index and MIPI.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Plateformes de Ressources Biologiques: PRB Créteil (PRB BB-0033-00021; Dijon, Lyon, Nancy, and IGR) and Dr N. Grardel and Prof de Leval who provided DNA from frozen tumor material. The authors also thank Isabelle Texier and Tony Noel from the Creteil laboratory for technical performance and Catherine Bely for secretarial assistance.
This work was supported by Institut National du Cancer (INCA), grant Programmes d'actions intégrées de recherche (PAIR Lymphome) 2008-1-LYM-01 (O.H.), Association pour la recherche sur le cancer (ARC) (subvention 3730) (M.-H.D.-L.), Fondation de France grant 2004004029 (M.-H.D.-L.), Association pour la recherche thérapeutique génétique et immunologique dans les lymphomes (ARTGIL) (C.H. and M.-H.D.-L.), Lymphoma Research Foundation (LRF) (M.D.), Framework Programmes 6 European Commission (contract LSHC-CT-2004-503351) (M.D.), and Roche (support for the MCL Younger trial) (M.D.). This work was supported in part by MRC Holland, which provided free kits for MLPA technology development.
Authorship
Contributions: M.-H.D.-L. designed the research, performed genetic experiments, and drafted the manuscript; W.K., F.B., and J.B. performed pathologic review and Ki-67 counts; E.M., C.P., and M.-H.D.-L. collected and characterized tumor infiltration of diagnostic samples during the clinical trial; G.S., O.C., P.F., C.H., V.R., C.T., and M.D. included studied patients in the clinical trial, helped in tumor tissue recovery, and, with F.J. and E.M., edited the manuscript; E.H. planned and performed the statistical analysis; O.H., M.U., and M.D. coordinated the MCL Younger trial; and all authors reviewed and accepted the final version of the manuscript. A list of the European mantle cell network members is available on http://www.european-mcl.net/en/members.php.
Conflict-of-interest disclosure: M.-H.D.-L., W.K., F.B., F.J., J.B., O.C., V.R., M.U., E.M., C.P., and O.H. declare no competing financial interests. E.H. received travel support by Roche. P.F. and C.H. received honoraria for advisory board participations from Roche. G.S. received research support and honoraria for conferences or advisory board participations from Roche/Genentech and honoraria for conferences or advisory board participations from Celgene and Janssen. M.D. received speaker’s honoraria and support of investigator-initiated trials (institution) from Roche.
Correspondence: Marie-Helene Delfau-Larue, Département d’Hématologie et Immunologie Biologiques, GH Mondor, AP-HP, INSERM U955 eq9, 94000 Créteil, France; e-mail: marie-helene.delfau@hmn.aphp.fr.