Abstract
Introduction
This study addresses the unsolved clinical problem of relapse in MLL-rearranged infant (<1 year old) acute lymphoblastic leukemia (ALL): 80% of infants with ALL harbor MLL rearrangements (MLL-r) and 66% of these will relapse and die with current treatment [5-year event free survival is 34%]. The typical pattern of failure in MLL-r ALL is successful remission induction (>90%) followed by early relapse (60-80%), suggesting rapid emergence of one or more chemoresistant subclones. Recently, whole genome sequencing of 47 MLL-r infant ALL cases showed a mean of only 1.3 non-silent mutations in the predominant leukemic clone (Andersson Nat. Gen. 2015;47:330-7). The critical barrier to progress in identifying novel, more effective therapies is this striking paucity of genomic changes that are found in addition to the MLL rearrangement. Therefore we hypothesized that epigenetic changes drive evolution of chemoresistant subclones and cause relapse.
Methods
We tested this hypothesis on two MLL-r infant ALL cases that had paired diagnosis-relapse (DX-RL) leukemic specimens using whole genome bisulfite sequencing (WGBS) and RNA sequencing (RNA-seq). We prepared WGBS and RNA-seq libraries and performed paired-end sequencing (2x100bp) using Illumina HiSeq 2500. Paired-end reads received a methylated (1) or unmethylated (0) calls at each contiguous CpG they cover producing a data set of four 0/1-state-long "epialleles" with 16 possible methylation state patterns ("0-0-0-0" to "1-1-1-1"). For each epiallele, we calculated the frequency of each of the 16 patterns at DX and RL. We developed analysis methods to align bisulfite-converted reads to the genome, call methylation states at CpGs within reads, assess subclonal "epiallele" frequency changes between DX and RL, map the genomic locations of epialleles to promoter regions of their corresponding gene isoforms, and quantify differential gene isoform expression between DX and RL.
Results
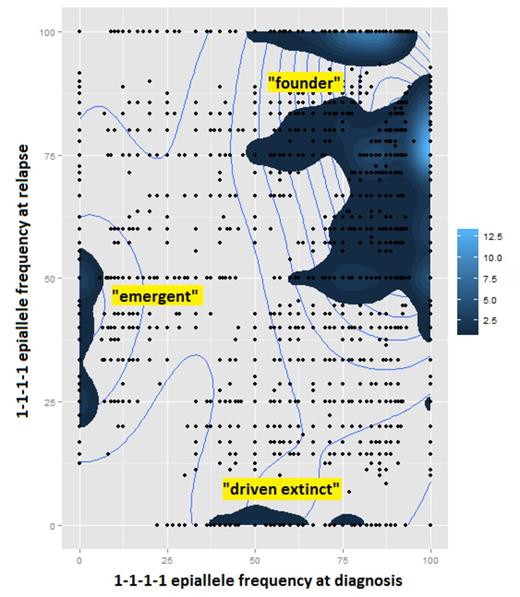
WGBS generated ~400 million paired-end reads per sample with 25X average coverage. After removing duplicates and repeats, we obtained methylation calls at ~15 million CpGs per case. RNA-seq generated RNA transcript quantification for ~47,000 isoforms from ~30,000 genes per sample. We extracted from the WGBS data paired-end reads that map uniquely to gene promoter regions and that cover at least 4 contiguous CpGs to a depth of at least 4X. In all, case A had 237,040 epialleles annotated to 11,484 gene promoters, and case B had 853,678 epialleles annotated to 14,717 gene promoters. By plotting the change in epiallele frequency between DX and RL, we were able to identify epiallele-defined subclones that were: not detectable at DX but present at RL ("emergent" at RL), present at DX but undetectable at RL ("driven extinct" at RL), and unchanged significantly between DX and RL ("founder") (Fig. 1). For example, in case A, we found 145 "1-1-1-1" epialleles that were at 0% frequency at DX and 66±17% (mean±st.dev.) frequency at RL (corresponding to 76±11% methylation increase at RL) that were associated with 29±77-fold decrease in expression of their associated gene isoform. 18 of these epialleles were: mapped to the promoter of IKZF1 (a known tumor-suppressor gene in ALL), present at 0% frequency at DX and 40±0.04% at RL, and associated with 8.5±10-fold decrease in IKZF1 isoforms expression. These results support the hypothesis that subclonal methylation changes associated with corresponding expression changes may drive chemoresistance and relapse.
Conclusions
We have developed analytic techniques to integrate WGBS and RNA-seq data for discovery of subclonal epigenetic changes that either emerge or are driven extinct under the selective pressure of chemotherapy treatment. Such data provide support for the hypothesis that chemoresistance in this disease may be driven by epigenetic mechanisms.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal