Abstract
Background
Shortened telomeres are seen in approximately a third of patients with idiopathic aplastic anemia (AA) and they also define risk of relapse, clonal evolution and overall survival. Constitutional pathogenic mutations in the telomere gene complex (TGC) are associated with very short telomeres (typically <1st centile) and may also present with AA, but have important implications in terms of different management strategies, especially with conditioning regimen and donor selection for bone marrow transplantation, which in turn affect transplant outcomes. Furthermore, detection of mutations of the TGC complex then entail regular screening/review of organ/systems affected in telomere disease, counselling and health insurance with regards to the increased risk of cancer and screening of family members. Novel variants affecting telomere function are increasingly being reported but in the absence of a full telomere disease phenotype, unaffected family members or variable penetrance of the mutant in affected family members, there remains uncertainty as to whether some are pathogenic or represent polymorphisms.
Methods
We report results using telomere length (TL) measurement by quantitative-real time PCR (qPCR) as a screening tool to identify patients for further mutation analysis on a customised panel of 10 TGC genes (TERT, TERC, DKC1, TINF2, NHP2, NOP10, RTEL1, CTC1, USB1 and WRAP) using deeply parallel sequencing in a cohort of patients with AA. Using a Polyphen score that predicts a variant to be possibly pathogenic, we have used telomere repeat amplification protocol (TRAP) assay to increase the robustness of classifying a variant as more likely pathogenic, where family history was unrevealing. TRAP assay was performed by introducing the variants into W138V13 cell line and comparing telomerase expression in them to wild type TERT and TERC expression of telomerase.
Results
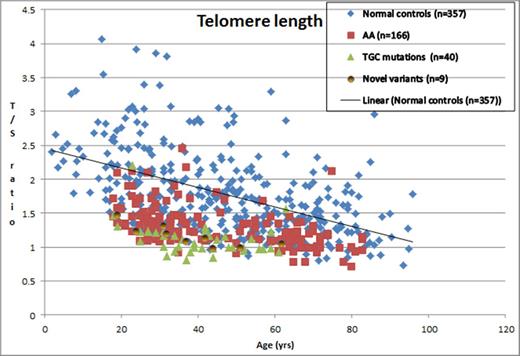
From the King's College Hospital database, we screened 295 patients with AA for TL using qPCR. The median age of the cohort was 44.2 years (range18.2- 83.4) with male/female ratio of 57:43. 189 patients (64%) had TL < 10th centile and 111 (37.6%) had TL <1st centile (Figure 1). We screened 215 of these patients for TGC mutation analysis and report 40 mutations (18.6%) in this cohort. Most mutations were in the reverse transcriptase enzyme TERT (n=33) and the remaining in the catalytic unit of the RNA complex TERC (n=7). A positive family history of bone marrow failure was seen in only 4 (10%) of these cases with TGC mutation, where the same TGC mutation was detected. 38/40 (95%) patients with a TGC mutation had a TL <10th centile and 32 (80%) had TL<1st centile. 32/111 (28.8%) of AA patients with TL<1st centile were found to have a TGC mutation, while only 6/78 (7.8%) of AA patients with 10th < TLth centile had TGC mutations.
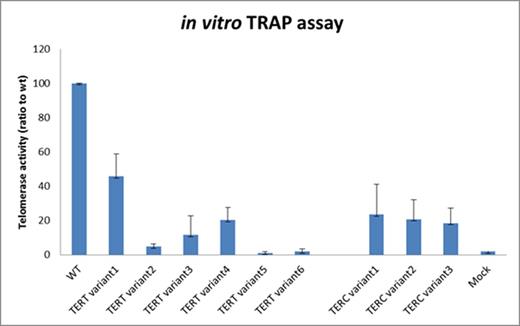
We identified a further 9 novel variants which were predicted to be possibly pathogenic, but have not been reported pathogenic in literature, as yet. All 9 patients with the novel variants had TL <1st centile with a median age of 37 (range 19-62), but had no positive family history of bone marrow failure or features of telomere disease. Using the TRAP assay, all 9 variants had telomerase activity well below 50% of WT TERT and TERC telomerase expression. All TERC variants had TRAP score of ~20% and 5 of the 6 TERT variants had TRAP score of <30% of WT TERT telomerase activity, of which 3 had telomerase activity <5% (Figure 2).
Conclusion
Two thirds of patients with AA have TL <10 th centile. Most patients with a TGC mutation have TL <1st centile. TL can reliably be used as a screening tool to investigate patients for further TGC mutation analysis. Heterozygous state mutation in TERT are the commonest, followed by TERC and explain the slightly late onset (mid 40's) and milder presentation of telomere disease as compared to the more severe phenotype and younger presentation classically seen with the homozygous state DKC mutations. A telomere length and a TRAP assay can add value in predicting a possible or putative pathogenic variant in a TERT or TERC gene on TGC analysis, where a reliable family history of telomereopathy is lacking.
Telomere length expressed as T/S ratio against age for the different cohorts
Telomere length expressed as T/S ratio against age for the different cohorts
Expression of telomerase activity in novel TERT and TERC variants compared to Wild-type (positive control) and Mock (negative control)
Expression of telomerase activity in novel TERT and TERC variants compared to Wild-type (positive control) and Mock (negative control)
Kulasekararaj:Alexion: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal