Abstract
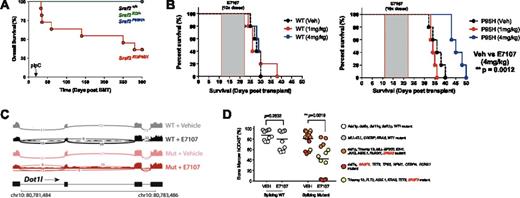
Mutations in the spliceosomal genes SRSF2, U2AF1 and SF3B1 are commonly found in patients with myeloid malignancies including myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). These mutations occur at highly restricted amino acid residues, are always heterozygous, and rarely co-occur with one another. These data suggest that splicing factor mutations confer an alteration of splicing function and/or that cells may only tolerate a certain degree of splicing modulation. Consistent with this, it has been recently shown that mice expressing the heterozygous Srsf2P95H mutation develop MDS-like features due to altered mRNA recognition. To begin to identify therapeutic vulnerabilities created by spliceosomal gene mutations, we first generated Mx1-Cre Srsf2P95H/KO mice to study the effects of wildtype Srsf2 deletion with concomitant activation of the Srsf2P95H mutant allele. In noncompetitive transplantation, hemizygous Mx1-Cre Srsf2P95H/KO mutant mice exhibited severe bone marrow failure due to complete loss of hematopoietic stem/progenitor cells (HSPCs), suggesting that Srsf2P95H cells depend on wildtype Srsf2 for survival (Fig. 1A). RNA-seq analyses revealed an increase in mis-splicing due to altered mRNA recognition in Srsf2P95H/KO hemizygous mutant HSPCs compared with heterozygous Srsf2P95H/+ mice, which resulted in strong repression of genes involved in hematopoietic self-renewal, most notably Runx1, Erg, and the entire HoxA cluster.
Based on these observations, we hypothesized that spliceosomal-mutant leukemias might display greater sensitivity to pharmacologic modulation of splicing than their wildtype counterparts. To address this, we tested the efficacy of a known spliceosome modulator, E7107, in myeloid leukemias with or without splicing factor mutations in vivo. We created a murine leukemia model using MLL-AF9 overexpression on either Srsf2+/+ or Srsf2P95H/+ background, a mutational combination which represents ~10% of adult MLL-rearranged leukemias (LavallŽe et al., 2015). In vivo E7107 treatment resulted in decreased disease burden and ultimately survival benefit in Srsf2P95H/+ mice, whereas the same treatment regimen had no impact on overall survival of Srsf2+/+ mice (Fig. 1B). RNA-seq analysis of leukemic cells after 5 consecutive days ofE7107 treatment in vivo revealed massive global splicing inhibition in response to E7107 in both genotypes, most notably global cassette exon skipping and retention of both constitutive and alternative introns. E7107-induced splicing inhibition was more severe in Srsf2P95H/+ versus Srsf2+/+ mice, consistent with its differential effect on survival in these 2 genotypes. Interestingly, among the most differentially spliced genes in Srsf2P95H/+ mice in response to E7107 are genes involved in maintaining the leukemogenic programs in MLL-rearranged leukemias, including Dot1l, Meis1 and Lsd1 (Fig. 1C).
To confirm that preferential sensitivity of spliceosomal-mutant myeloid leukemias to splicing modulation is directly translatable to a human context, we next generated patient-derived xenografts (PDX) from AML patients wildtype (n=2) or mutant (n=3) for splicing factors. 10 mice were successfully engrafted per patient via tail-vein injection in NSG mice and treated with vehicle or E7107 for 10 days once human bone marrow chimerism was >50%. In each PDX model, targeted genomic analysis of human leukemic cells confirmed faithful engraftment of the major leukemic clones as found in patients pre-engraftment and that spliceosomal mutations were present in the major leukemic clone engrafted in mice. All spliceosomal mutant AML PDXs showed significant reductions in human leukemic burden in response to E7107 while the response in splicing wildtype AML cells was less robust (Fig. 1D).
Collectively, these data provide both genetic and pharmacologic evidence that splicing factor mutant leukemias are preferentially sensitive to splicing modulation in vivo compared with spliceosomal wildtype counterpart cells. This has important therapeutic implications for the wide range of patients with MDS and AML bearing these alterations.
Thomas:H3 Biomedicine: Employment. Buonamici:H3 Biomedicine: Employment. Smith:H3 Biomedicine Inc.: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal