Abstract
Molecular mutations are routinely incorporated into the risk stratification of newly diagnosed acute myeloid leukemia (AML) patients (pts). However, their prognostic impact in relapsed AML has not been well characterized. To better understand the landscape and prognostic impact of molecular mutations in relapsed AML, we retrospectively examined pts with AML in first relapse.
Methods: We performed multi-amplicon targeted deep sequencing on samples from bone marrow and peripheral blood of pts diagnosed with AML in first relapse at our institution between the years 2002-2013. A multiple gene sequencing panel of 64 genes that have been described as recurrently mutated in myeloid malignancies was applied. Genes were analyzed individually and grouped based on their functional pathways: RNA splicing, RNA-helicase family, DNA methylation/epigenetic, transcription factors, signal transduction/ receptors, histone/ chromatin modification, cohesion complex. Cytogenetic (CG) risk was ascribed by CALGB/Alliance 8461 criteria. The primary endpoints were response to salvage therapy and overall survival from relapse (OS-R). Binary logistic and proportional hazards models were used for univariable and multivariable analyses. Multivariable analyses of individual genes and functional groups used a stepwise variable selection algorithm and were based on the genes and gene groups found to be significant in univariable analysis at p < 0.20.
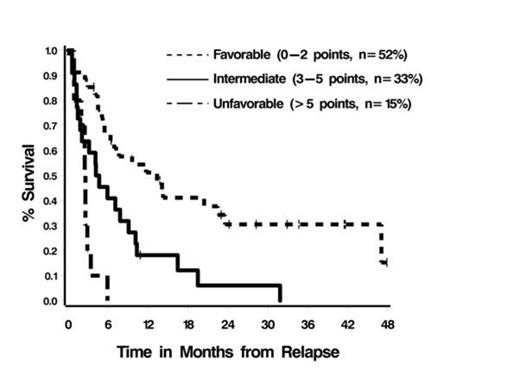
Results: Data from 74 pts were analyzed. The median age at diagnosis was 61 years (range 27-77). CG risk at diagnosis: good (17%), intermediate (42%), miscellaneous (15%), poor (26%). At the time of relapse, 68% of pts received intensive salvage chemotherapy [i.e. MEC (mitoxantrone, etoposide, cytarabine), high dose cytarabine], 14% supportive care, and 19% low dose therapy. Sixteen percent of pts received an allogeneic hematopoietic stem cell transplant. The most common mutations were found in NPM1 (16%), DNTM3A (15%), IDH2 (14%), and PHF6 (10%). Overall, 19 pts (26%) had no mutations, 25 (34%) had 1, 12 (16%) had 2, 11 (15%) had 3, and 7 (9%) had > 3 mutations. Fifty-two percent of pts achieved a complete remission (CR) or CR with incomplete count recovery (CRi) with salvage therapy (95% CI: 39-64%) and median OS-R was 5.2 months. In multivariable analyses, the number of mutations (0, 1, >1) in DNMT3A, TP53, NRAS, and ASXL1 (panel 1) was the only factor found to be independently associated with worse OS-R and response to salvage therapy (p< 0.0001 and 0.03, respectively). Adjusting for age, CG risk, and treatment, the number of mutations in panel 1 remained a significant predictor of OS-R (p=0.003) but not of response to salvage therapy (p=0.23). Multivariable analyses of panel 1 and conventional CG risk suggested that both factors contained prognostic information (p < 0.0001 and 0.07, respectively); and that a modified CG risk classification system could be defined using a simple scoring algorithm that assigns points to each conventional CG risk group and the number of panel 1 mutations that are proportional to the corresponding regression coefficients from the model and then summing the number of points present (Table 1). Based on the scoring system, 3 prognostic groups can be defined (Figure 1).
Scoring Algorithm
| Factor . | Points . |
|---|---|
| CG Risk | |
| Good | 0 |
| Intermediate | 2 |
| Miscellaneous | 2 |
| Poor | 3 |
| Number of mutations in panel 1 | |
| 0 | 0 |
| 1 | 3 |
| >1 | 6 |
| Factor . | Points . |
|---|---|
| CG Risk | |
| Good | 0 |
| Intermediate | 2 |
| Miscellaneous | 2 |
| Poor | 3 |
| Number of mutations in panel 1 | |
| 0 | 0 |
| 1 | 3 |
| >1 | 6 |
Conclusions: Molecular mutations are prognostic for OS-R and may help identify patients who would benefit from novel therapies at the time of relapse. Specific predictive mutations included genes related to DNA methylation and histone modification suggesting that targeting these specific factors may improve outcome.
Carew:Boehringer Ingelheim: Research Funding. Sekeres:Celgene Corporation: Membership on an entity's Board of Directors or advisory committees; TetraLogic: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal