Abstract
Background:TET2 is a hematopoietic tumor suppressor gene that has been implicated in DNA demethylation and the epigenetic regulation of gene expression. Inactivating TET2 mutations are common in aging-associated clonal hematopoiesis of indeterminate potential (CHIP) and myelodysplastic syndromes (MDS). TET2 mutations may contribute to early clonal dominance and myeloid transformation, although the exact mechanisms remain to be elucidated. Emerging evidence suggests that the MDS bone marrow niche may be abnormal and this abnormal niche may act as fertile ground for expansion of neoplastic cells in vivo. Common to the environment of MDS and “inflammaging” are elevations in cytokines, such as TNFa and IFNg. We hypothesized that TET2 mutant clones may thrive in an inflammatory environment and further condition this environment to promote their own survival.
Methods: Adult (10-14 weeks-old) Tet2 wild type and Tet2 mutant C57BL/6 mice strains (JAX) were chosen as a model system. The floxed Tet2 allele was deleted by targeting exon 3 with Vav1-cre mediated, hematopoietic-specific excision. We isolated lineage negative cells (Lin-), enriched for hematopoietic stem and progenitor cells (HSPC), from Tet2 wild type and -/- and bone marrow (BM) (EasySep; StemCell Technologies) and cultured these in the absence or presence of TNFα (0.1, 1, or 10 ng/ml) and IFN-γ (0.01, 0.1 or 1 ng/ml) in a methylcellulose colony formation assay (MethoCult; StemCell) or liquid culture media, and then examined their colony growth, cell count and phenotypic characterization over a period of 12 days. Where indicated, serial re-plating was performed.
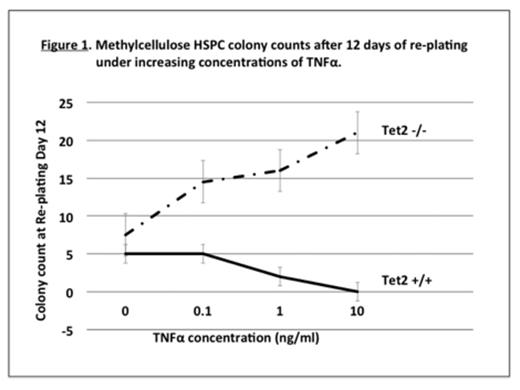
Results: We found an increased proportion of Lin- cells in Tet2 -/- BM compared to wild type, suggesting in vivo HSPC expansion. In triplicate experiments starting with equal numbers of wild type and Tet2 -/- Lin- cells (104 cells/MethoCult well), we found no significant difference in colony counts on days 3, 6, 9, or 12, when cultured in the absence or presence of increasing TNFα concentrations. As expected, TNFα dose-dependently reduced colony counts in both genotypes (up to 3 to 4-fold at 10 ng/ml). However, Tet2 -/- Lin- cells displayed a proliferative advantage over wild type in serial re-plating assays. In the presence of TNFα, this Tet2 -/- re-plating advantage was striking. As exemplified by day 12 colony counts at first re-plating (Fig. 1), while wild type colonies declined with increasing TNFα, Tet2 -/- colony counts increased with TNFα concentration (i.e. average 20-fold higher than wild type at 10 ng/ml TNFα; p<0.05).
We next shifted our analysis to IFNg, and found significantly increased day 6, 9 and 12 Tet2 -/- methylcellulose colonies at first plating. Upon re-plating in IFNg, Tet2 -/- cells demonstrated significantly increased (1.5 to 2-fold; p<0.05) mean day 12 colony counts at 0.01, 0.1 and 1 ng/ml IFNg. To gain some insight into the nature of the cells emerging under IFNg stress, we performed flow cytometry upon re-plating. Preliminary experiments revealed increases (1.5 to 2-fold) in Mac1+Gr1+ and Sca1+Kit1+ populations in Tet2 -/-, as compared to wild type, in the presence of IFNg.
We are currently comparing apoptosis in wild type and Tet2 -/- cells in the MethoCult system +/- TNFα and IFNg (and a more amenable liquid culture system), using Annexin V/Propidium Iodide-based flow cytometry. These results will be reported. Future directions include the characterization of differential: a) gene expression signatures in Tet2 wild type and -/- Lin- cells under TNFα and IFNg stress, and b) TNFα and IFNg signatures in TET2 -mutant and non-mutant human MDS.
Conclusion:Tet2 -deficient murine bone marrow progenitors demonstrate a proliferative advantage, as compared to their wild type counterparts, under TNFα and IFNg stress. Given that these inflammatory cytokines have been associated with inflammaging and myelodysplasia, it is worth exploring whether TET2 -mutant human clones may emerge under inflammatory stress, leading to CHIP and/or MDS, and presenting a novel therapeutic target for clone eradication.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal