Abstract
Background: Cytogenetic and molecular markers, in addition to age and WBC count, at presentation have been the conventional standards for risk stratifying patients with ALL at diagnosis. There is increased recognition of the oligo-clonal nature of most acute leukemias and hence the limitation of molecular techniques at diagnosis in risk stratification.We hypothesized that the immunophenotype (IPT) heterogeneity as detected by multiparameter flow cytometry (MPFC) will be a function of the complex underlying genetic abnormality and better prognosticate B-ALL at diagnosis.
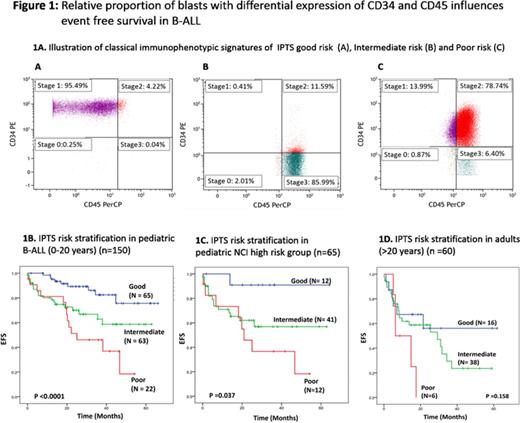
Methods: Retrospectively, the "raw" list mode files of 150 pediatric (0-20years) and 60 adult (>20 years) patients with B-ALL were analyzed (Jan 2009 - Dec 2013). All patients were diagnosed and classified as per WHO 2008 criteria. On MPFC, four blast sub-populations were identified using CD34 and CD45 expression. CD45 expression was divided into bright and dim, using threshold set at one log below CD45 median fluorescence intensity (MFI) of residual lymphocytes while CD34+ population was identified using unstained cells and known internal negative controls (lymphocytes). Four sub-populations were identified and named based upon resemblance to stages of hematogones - Stage 1: CD34+CD45dim, Stage 2: CD34+CD45bright, Stage 3: CD34-CD45bright and stage 0: CD34-CD45dim (Illustrated in Fig 1A). Conventional chemotherapy schedule as previously reported by us was used to treat the patients (Bajel et al. Leukemia:2007).
Results: As a continuous variable, proportion of blasts in stage 1, 2, 3, but not stage 0, correlated with events (relapse or death). A high proportion of blasts in Stage 1 had positive impact while high proportion of Stage 2 and 3 had negative impact on EFS. Using uni-variate Cox regression analysis, we tested different cut-offs for all three stages and each sub-population was scored proportional to the hazard ratio as follows: Stage 1 ≤50% = score 2.5, Stage 2 ≥25%: score 2.5 and Stage 3 ≥20%: score 2. All cases were then scored for each parameter and a combined immunophenotype score (IPTS) was calculated. Each patient could have a score varying from 0 to 7. After clustering the KM analysis curves, patients were classified as good risk if the IPTS was 0, high risk if IPTS was >4.5 and the rest were intermediate IPTS. There were significant differences in the clinico-genetic and survival profiles of three categories which are summarized in Table 1 and Figure 1B. On multivariate Cox regression analysis for occurrence of event (including age, high WBC count, BCR-ABL1, cytogenetic risk stratification, BFM-95 risk stratification and post-induction status) IPTS poor risk emerged as an independent prognostic variable (HR:4.7, 95%CI: 1.744-12.697;p=0.002). In adult patients (n=60) similar trends were seen, although, differences were not statistically significant due to small number of patients in which it was evaluated (Figure 1D).
Conclusion: Patients with different immunophenotypic composition at diagnosis are associated with distinct clinical profile, genetic features and treatment outcomes. Alone or in combination with a limited conventional molecular profile, this could serve as a low cost but effective strategy to risk stratify patients at diagnosis, especially in a resource limited setting.
Comparison of clinical and genetics features of IPTS risk groups in pediatric cohort.
| . | Good (n=65) N (%), Mean ±SD . | Intermediate (n=63) N (%), Mean ±SD . | Poor (n=22) N (%), Mean ±SD . | P value . |
|---|---|---|---|---|
| Age (years) | 6.0 ±4.2 | 11.1 ±5.8 | 9 ±6.5 | 0.000 |
| WBC count (x109/L) | 11.5 ±15.2 | 37.6 ±76.4 | 45.8 ±114.7 | 0.045 |
| BCR-ABL1 | 0(0) | 4(6.3) | 3(13.6) | 0.023 |
| ETV6-RUNX1 | 10(15.4) | 5(7.9) | 1(4.5) | 0.246 |
| TCF3-PBX1 | 0(0.0) | 7(11.1) | 0(0.0) | 0.018 |
| Hyperdiploidy | 39(60.0) | 13(20.63) | 5(22.7) | 0.000 |
| NCI risk stratification | 0.000 | |||
| Standard | 53(81.5) | 22(34.9) | 10(45.4) | |
| High | 12(18.5) | 41(65.1) | 12(54.5) | |
| Event | 9(13.8) | 20(31.7) | 13(59.1) | 0.000 |
| Relapse | 9(13.8) | 12(19.05) | 12(54.5) | 0.000 |
| Death | 7(10.8) | 17(27.0) | 9(40.9) | 0.006 |
| . | Good (n=65) N (%), Mean ±SD . | Intermediate (n=63) N (%), Mean ±SD . | Poor (n=22) N (%), Mean ±SD . | P value . |
|---|---|---|---|---|
| Age (years) | 6.0 ±4.2 | 11.1 ±5.8 | 9 ±6.5 | 0.000 |
| WBC count (x109/L) | 11.5 ±15.2 | 37.6 ±76.4 | 45.8 ±114.7 | 0.045 |
| BCR-ABL1 | 0(0) | 4(6.3) | 3(13.6) | 0.023 |
| ETV6-RUNX1 | 10(15.4) | 5(7.9) | 1(4.5) | 0.246 |
| TCF3-PBX1 | 0(0.0) | 7(11.1) | 0(0.0) | 0.018 |
| Hyperdiploidy | 39(60.0) | 13(20.63) | 5(22.7) | 0.000 |
| NCI risk stratification | 0.000 | |||
| Standard | 53(81.5) | 22(34.9) | 10(45.4) | |
| High | 12(18.5) | 41(65.1) | 12(54.5) | |
| Event | 9(13.8) | 20(31.7) | 13(59.1) | 0.000 |
| Relapse | 9(13.8) | 12(19.05) | 12(54.5) | 0.000 |
| Death | 7(10.8) | 17(27.0) | 9(40.9) | 0.006 |
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal