Abstract
Mutations in genes encoding the isocitrate dehydrogenase 1/2 (IDH 1/2) enzymes increase production of the oncometabolite 2-hydroxyglutarate (2HG), resulting in elevated 2HG in patients with IDH-mutant AML. This may allow for non-invasive diagnostic and predictive markers of disease; however, the optimal threshold of 2HG levels to predict IDH mutation status, or whether 2-HG measurements in different compartments are equally predictive, is unknown. We measured 2-HG levels in serum, urine, bone marrow aspirate, and bone marrow cell pellet to determine the optimal predictive value of 2HG levels for IDH mutations at AML diagnosis.
Patients with newly diagnosed AML had prospective measurements of 2HG levels at diagnosis by liquid chromatography-tandem mass spectrometry in serum, urine, marrow aspirate, and marrow pellet samples. Patients analyzed had IDH1 R132 and IDH2 R140 and R172 testing. Hotspot mutational profiling was performed for AKT1, APC, BRAF, CTNNB1, EGFR, ERBB2, KIT, KRAS, MAP2K1, NOTCH1, NRAS, PIK3CA, P53, and PTEN; patients were also tested for FLT3, NPM1, and CEBPA mutations. IDH1/2 mutant patients were compared to wildtype (WT) patients using a Wilcoxon rank sum, Fisher's exact, or Kruskal Wallis test, as appropriate. Performance characteristics of 2HG to predict the presence of IDH1/2 mutations were done using a recursive partitioning algorithm in R version 3.2.1, with the rpart package.
228 patients with newly diagnosed AML had 2HG levels in serum, urine, marrow aspirate, and marrow pellet samples. All patients had testing for IDH1 R132 and IDH2 R140 and R172 mutations. The patients were 56% male; 13 patients had APL, none with IDH mutations. IDH mutations were identified in 23% (n=52) of the cohort: IDH1 R132C (n=12), IDH2 R172 (n=9), and IDH2 R140Q (n=29), and 2 additional patients had mutations in both IDH1 R132 and IDH2 R140 (Table 1).
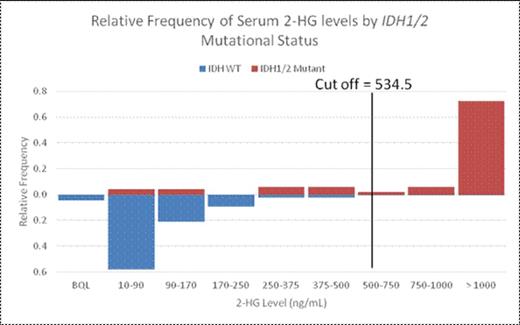
The optimal test cut-off of 2HG to predict IDH status was 534.5 ng/mL in the serum (n=221), 16650 ng/mL in the urine (n=213), 2210 ng/mL in the marrow aspirate (n=190), and 1146 ng/2*10^6 cells in the pellet (n=159). Serum and pellet values had the greatest specificity for the presence of an IDH1/2 mutation (0.9882 and 1.000, respectively; Table 2). The positive predictive value of an elevated serum or pellet 2HG level at these cut-offs was 95.4% and 100%, given a 23% IDH mutation prevalence in this study population. The marrow aspirate had the greatest sensitivity (0.8837) and negative predictive value (96.6%). Urine 2HG levels were less sensitive than serum 2HG levels, although above the urine cut-off the specificity for IDH mutations was similar (Figure 1).
2HG levels in the serum, urine, and marrow can be used to identify IDH mutations in AML. Serum 2HG testing is an effective non-invasive mechanism to predict IDH1/2 mutation status. A serum cut-off of 534.5 ng/mL has a specificity of 0.9882 and, with an IDH mutation prevalence of 23%, was associated with a PPV of 0.9535.
Patient Characteristics
| . | No IDH mutation . | IDH mutation . | Total . | p-value . |
|---|---|---|---|---|
| Male sex | 101 (57%) | 26 (50%) | 127 (56%) | 0.43 |
| Cytogenetics | 0.0002 | |||
| Favorable | 26 (15%) | 0 | 26 (12%) | |
| Intermediate | 95 (54%) | 41 (82%) | 136 (61%) | |
| Adverse | 52 (30%) | 9 (18%) | 61 (27%) | |
| Age (median, range) | 66 (20, 87) | 66.5 (41, 86) | 66 (20, 87) | 0.27 |
| WBC (median, range) | 5.35 (0.60, 315.4) | 3.65 (0.20, 333.2) | 5.25 (0.20, 333.2) | 0.33 |
| No IDH mutation | IDH mutation | Number assessed | p-value | |
| NRAS | 27 (17%) | 9 (19%) | 210 | 0.83 |
| KRAS | 16 (10%) | 0 (0%) | 210 | 0.03 |
| TP53 | 15 (9%) | 1 (2%) | 210 | 0.13 |
| KIT | 2 (1%) | 1 (2%) | 210 | 0.54 |
| FLT3ITD | 34 (20%) | 4 (8%) | 218 | 0.06 |
| FLT3TKD | 10 (6%) | 2(4%) | 218 | 0.74 |
| NPM1 | 29 (17%) | 15 (30%) | 219 | 0.07 |
| CEBPA | 13 (13%) | 3 (10%) | 102 | 1.00 |
| 2HG Measurements | No IDH mutation | IDH mutation | Total | |
| Serum (ng/mL) | 79.5 [52,123] n=170 | 1420 [675,2735] n=51 | 101 [58,101] n=221 | |
| Urine (ng/mL) | 3590 [2230,6220] n=163 | 18300 [7260,59500] n=50 | 4330 [2450,8580] n=213 | |
| Marrow aspirate (ng/mL) | BQL [BQL,BQL] n=147 | 18400 [4270,43100] n=43 | BQL [BQL,1430] n=190 | |
| Pellet (1000 ng/2*10^6 cells) | 64 [BQL,169] n=127 | 1420 [675,2735] N=32 | 107 [BQL,500] n=159 |
| . | No IDH mutation . | IDH mutation . | Total . | p-value . |
|---|---|---|---|---|
| Male sex | 101 (57%) | 26 (50%) | 127 (56%) | 0.43 |
| Cytogenetics | 0.0002 | |||
| Favorable | 26 (15%) | 0 | 26 (12%) | |
| Intermediate | 95 (54%) | 41 (82%) | 136 (61%) | |
| Adverse | 52 (30%) | 9 (18%) | 61 (27%) | |
| Age (median, range) | 66 (20, 87) | 66.5 (41, 86) | 66 (20, 87) | 0.27 |
| WBC (median, range) | 5.35 (0.60, 315.4) | 3.65 (0.20, 333.2) | 5.25 (0.20, 333.2) | 0.33 |
| No IDH mutation | IDH mutation | Number assessed | p-value | |
| NRAS | 27 (17%) | 9 (19%) | 210 | 0.83 |
| KRAS | 16 (10%) | 0 (0%) | 210 | 0.03 |
| TP53 | 15 (9%) | 1 (2%) | 210 | 0.13 |
| KIT | 2 (1%) | 1 (2%) | 210 | 0.54 |
| FLT3ITD | 34 (20%) | 4 (8%) | 218 | 0.06 |
| FLT3TKD | 10 (6%) | 2(4%) | 218 | 0.74 |
| NPM1 | 29 (17%) | 15 (30%) | 219 | 0.07 |
| CEBPA | 13 (13%) | 3 (10%) | 102 | 1.00 |
| 2HG Measurements | No IDH mutation | IDH mutation | Total | |
| Serum (ng/mL) | 79.5 [52,123] n=170 | 1420 [675,2735] n=51 | 101 [58,101] n=221 | |
| Urine (ng/mL) | 3590 [2230,6220] n=163 | 18300 [7260,59500] n=50 | 4330 [2450,8580] n=213 | |
| Marrow aspirate (ng/mL) | BQL [BQL,BQL] n=147 | 18400 [4270,43100] n=43 | BQL [BQL,1430] n=190 | |
| Pellet (1000 ng/2*10^6 cells) | 64 [BQL,169] n=127 | 1420 [675,2735] N=32 | 107 [BQL,500] n=159 |
Test characteristics based on optimal 2HG cut-off, by compartment.
| Compartment . | 2HG cut-off . | Sensitivity . | Specificity . | . | 23% prevalence . | |
|---|---|---|---|---|---|---|
| . | . | . | . | . | PPV . | NPV . |
| Serum | 534.5 ng/mL | 0.8039 | 0.9882 | 0.9535 | 0.9438 | |
| Urine | 16650 ng/mL | 0.5600 | 0.9877 | 0.9333 | 0.8798 | |
| Marrow Aspirate | 2210 ng/mL | 0.8837 | 0.9660 | 0.8837 | 0.9660 | |
| Marrow Pellet | 1146 ng/2*10^6 cells | 0.7813 | 1.000 | 1.000 | 0.9478 | |
| Compartment . | 2HG cut-off . | Sensitivity . | Specificity . | . | 23% prevalence . | |
|---|---|---|---|---|---|---|
| . | . | . | . | . | PPV . | NPV . |
| Serum | 534.5 ng/mL | 0.8039 | 0.9882 | 0.9535 | 0.9438 | |
| Urine | 16650 ng/mL | 0.5600 | 0.9877 | 0.9333 | 0.8798 | |
| Marrow Aspirate | 2210 ng/mL | 0.8837 | 0.9660 | 0.8837 | 0.9660 | |
| Marrow Pellet | 1146 ng/2*10^6 cells | 0.7813 | 1.000 | 1.000 | 0.9478 | |
Relative frequencies of 2-HG levels according to cut-off values in serum (top) urine (bottom).
Relative frequencies of 2-HG levels according to cut-off values in serum (top) urine (bottom).
Chen:Bayer: Consultancy, Research Funding. Stone:Agios: Consultancy; Celator: Consultancy; Pfizer: Consultancy; Merck: Consultancy; AROG: Consultancy; Abbvie: Consultancy; Celgene: Consultancy; Roche/Genetech: Consultancy; Karyopharm: Consultancy; Amgen: Consultancy; Sunesis: Consultancy, Other: DSMB for clinical trial; Juno: Consultancy; Novartis: Research Funding. Fathi:Exelexis: Research Funding; Agios: Membership on an entity's Board of Directors or advisory committees; Seattle Genetics: Membership on an entity's Board of Directors or advisory committees, Research Funding; Merck: Membership on an entity's Board of Directors or advisory committees; Ariad: Consultancy; Takeda Pharmaceuticals International Co.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal