Abstract
Characterisation of antigen expression patterns is part of the standard diagnostic work-up in acute myeloid leukemia (AML). But the biological & clinical implication of such antigen expression patterns have not been studied extensively & remain unclear in AML patients (pts) undergoing allogeneic stem cell transplantation (SCT).
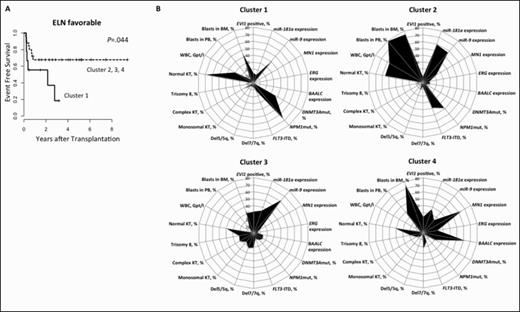
We analyzed the diagnostic antigen expression patterns of 162 AML pts (median age 64.7 years [y, range 46.6-76.2y]) with available data who received allogeneic peripheral blood SCT after non-myeloablative conditioning (NMA-SCT) between 2001 & 2013 at our institution. Conditioning regimen was fludarabine 3x30mg/m2 & 2Gy total body irradiation. Donors were human leukocyte antigen (HLA) matched related (12%) or HLA matched (59%) or mismatched unrelated (29%). Mutation (mut) status of the NPM1, CEBPA, IDH1, IDH2 & DNMT3A gene, presence of FLT3 -ITD & FLT3-TKD & the expression status of BAALC, ERG, MN1, EVI1, miR-9 & miR-181a at diagnosis were accessed. Pts were grouped according to the European LeukemiaNet (ELN) genetic classification in 22% favorable (fav), 24% intermediate-I (int-I), 21% intermediate-II (int-II) & 32% adverse (adv). Median follow up was 3.2y. To assess antigen expression patterns at diagnosis for all pts, flow cytometric analysis utilizing a standard panel (CD2, CD7, CD11b, CD13, CD14, CD15, CD33, CD34, CD38, CD45, CD56, CD61, CD64, CD65, CD117 & Glycophorin A) of mononuclear cells in bone marrow (BM) was performed. Using R's gplot package we performed unsupervised hierarchical clustering of the antigen expression which revealed 4 subgroups with distinct antigen expression patterns (Figure 1).
At diagnosis, pts grouped in cluster 1 (n=19) had higher white blood count (WBC, P=.004) & lower peripheral blood (PB) blast count (P =.03) & were more likely to have de novo AML (P =.05). They were also less likely to have trisomy 8 (P=.08) by trend & more likely to have normal karyotype (KT, P=.05), to have ELN fav risk (P =.04), to be NPM1 mut (P =.002) & to be DNMT3A mut by trend (P=.08) & had lower miR-181a (P=.04), lower BAALC (P<.001), lower ERG (P=.01) & lower MN1 expression (P<.001).
Pts grouped in cluster 2 (n=35) had higher WBC (P<.001), PB blasts (P<.001) & BM blasts (P=.005) at diagnosis. They were less likely to have trisomy 8 (P=.008) & to have deletion (del) 7/7q (P =.07) by trend, were more likely to be NPM1 mut (P =.002) & to have FLT3 -ITD (P <.001) & had lower BAALC (P =.1) & lower EVI1 expression (P =.09) by trend.
Pts grouped in cluster 3 (n=59) had lower WBC (P<.001), PB blasts (P<.001) & BM blasts (P<.001) at diagnosis & were less likely to have de novo AML (P<.001). They were more likely to have trisomy 8 (P=.05), del5/5q (P=.004), monosomal KT (P=.04), complex KT (P=.07) by trend & ELN adv risk (P=.04), were less likely to be NPM1 mut (P =.03) & FLT3 -ITD by trend (P=.08) & had lower ERG (P=.008) & higher miR-9 (P=.009) expression.
Pts grouped in cluster 4 (n=49) had lower WBC (P=.03), higher PB blasts (P=.007) & BM blasts (P<.001) at diagnosis. They were less likely to have del5/5q (P=.008) & NPM1 mut (P <.001) & had lower miR-9 (P=.007) & higher BAALC (P<.001), ERG (P<.001) & MN1 (P<.001) expression.
For the entire set of pts, belonging to one of the antigen expression clusters did not impact on outcome. However, when the ELN groups were regarded separately, within the ELN fav group, cluster 1 pts had a significantly shorter event free survival (EFS, P=.04, Figure 2A) & within the ELN int-I group, cluster 3 pts had a trend for better (P=.096) & cluster 4 pts for worse EFS (P=.087).
In conclusion, the antigen expression patterns at diagnosis obtained by unsupervised cluster analysis associated with distinct biological & clinical features (Figure 2B): NPM1 mut were enriched in clusters 1 & 2. Cluster 1 was characterized by ELN fav risk, normal KT, de novo disease & lower BAALC, ERG, MN1 & miR-181a expression. Cluster 2 was characterized by a high incidence of FLT3-ITD. We found more pts with ELN adv risk, monosomal KT, secondary AML & low miR-9 expression in cluster 3 & higher miR-9 as well as lower BAALC, ERG & MN1 expression levels in cluster 4. Even though we did not observe a prognostic impact of the antigen expression patterns in the entire cohort, the patterns may help to refine the ELN risk classification for AML pts undergoing SCT. Assessing the diagnostic antigen expression patterns provides information on disease biology, clinical parameters and potentially disease aggressiveness in AML.
Franke:BMS: Honoraria; MSD: Other: Travel Costs; Novartis: Other: Travel Costs. Niederwieser:Novartis: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal