Abstract
Background: Tumor heterogeneity has been documented in several malignancies and is generally associated with more aggressive disease and poorer prognosis. While heterogeneity has been documented in acute myeloid leukemia (AML), risk prognosis in AML is currently assessed by cytogenetics and the presence of recurrent mutations in genes such as FLT3 and NPM1, which are typically tested on a single gene basis. Prior approaches to identify molecular clonality have been laborious and therefore of limited clinical utility. The advent of next generation sequencing (NGS), however, allows for the rapid assessment of subclones within tumor populations. Currently, the prognostic significance of tumor heterogeneity in AML is not well defined. Therefore, this pilot study assessed the clinical significance of molecular clonality in AML using data generated by an NGS platform.
Methods: We selected patients with de novo AML who had blood or bone marrow submitted for sequencing at initial diagnosis. Patients who subsequently elected for comfort measures only without receiving AML-directed therapy were excluded. Study patients were sequenced with a 25 gene NGS panel on the Ion Torrent platform. This panel included genes that have been shown to be frequently mutated in AML and which lead to increased cellular proliferation or impaired differentiation, with particular emphasis on therapeutic and prognostic mutations. Germline mutations, identified by allelic fraction and/or minor allele frequency, were excluded. Remaining somatic mutations were analyzed with software employing a variational Bayesian algorithm to predict tumor heterogeneity such that patients were assessed for the presence of tumor heterogeneity (≥2 predicted clones) at the time of diagnosis. Heterogeneity was recorded and compared to several clinical parameters, including post-chemotherapy survival at 120 days, patient age, gender, karyotype, and association with the recurrent genetic mutations NPM1, FLT3-ITD, and CEBPA.
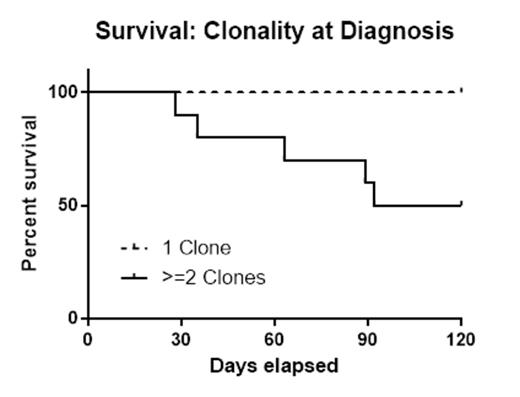
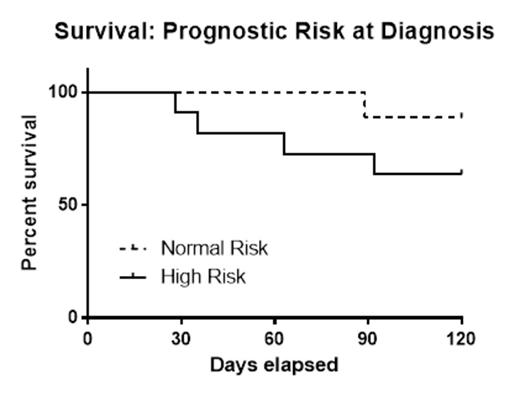
Results: Twenty patients met the study criteria of de novo AML and elected to receive therapy. The mean age at diagnosis was 59 years (38-77 years); overall survival was 75% at 120 days. Half of the patients (n=10) were determined to have ≥2 clones. Patients with either a complex karyotype or the FLT3-ITD mutation were classified as high risk (n=11). While survival at 120 days was lower in the high risk group (64%) compared to normal risk subjects (89%), this difference did not reach statistical significance (p=0.19). By contrast, patients with tumor heterogeneity (≥2 clones) had significantly lower 120 day survival (50%) when compared to AML patients with a single clone (100%) (p=0.01). There was no correlation between patients with multiple clones and high risk classification (p=0.18).
Conclusions: These pilot data present some of the first prospective evidence that heterogeneity detected by NGS is an adverse prognostic indicator for early survival in de novo AML post-chemotherapy. This finding may not be surprising given that the presence of tumor heterogeneity has been shown to confer adverse prognostic risk in several solid malignancies. Still, this early impact of molecular clonality in AML may have important consequences on choice of therapy. Further prospective studies are needed to confirm our findings and to determine the long-term outcomes in AML patient subsets, as well as examining whether a distinct approach to therapy in AML patients with clonal heterogeneity leads to improved early survival.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal