Abstract
Background: AML is a group of genetically distinct entities which have been defined by the presence of certain recurrent, mutually exclusive genetic abnormalities such as AML-specific fusion genes (e.g. PML-RARA, RUNX1-RUNX1T1) or recurrent molecular mutations (NPM1 mutations and CEBPA double mutations (dm)). In addition partial tandem duplications within the MLL gene (MLL-PTD) and RUNX1 mutations seem to play an important role in certain AML subtypes. A subset of AML with normal karyotype lacking the above-mentioned mutations is still poorly characterized. Array CGH and fluorescence in situ hybridization are able to detect abnormalities which are undetectable by chromosome banding analysis either due to a higher resolution, ability to detect copy neutral loss of heterozygosity (CN-LOH) and independence of in vitro proliferation.
Aims: 1. Search for submicroscopic copy number changes and cryptic rearrangements in AML with normal karyotype lacking NPM1 and RUNX1 mutations, CEBPA dm, and MLL-PTD. 2. Determine whether submicroscopic cytogenetic changes impact on survival.
Patients and Methods: For 1473 AML cases with normal karyotype complete data on mutation status of NPM1, CEBPA, RUNX1 and MLL -PTD was available. Of these 303 cases (21%) did not carry one of these mutations. Out of these 159 cases with de novo AML (median age: 68 years (range: 19-93)) were selected on the basis of availability of material for array CGH (SurePrint G3 ISCA CGH+SNP Microarray, Agilent, Waldbronn, Germany) and FISH screening with probes for MLL, RUNX1, CBFB, NUP98, MECOM/EVI1, NPM1, ETV6 and DEK-NUP214 (MetaSystems, Altlussheim, Germany; ABBOTT, Wiesbaden Germany).
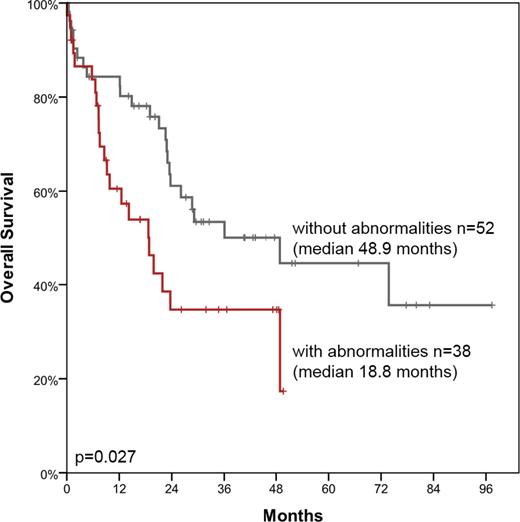
Results: In total in 67 of 159 patients (42%) abnormalities were identified by FISH and/or array CGH. In detail, 12 balanced rearrangements were detected by FISH screening involving NUP98 (n=7, in 6 of these a NUP98-NSD1 rearrangement was identified by PCR), MLL (n=2) and MECOM, RUNX1 and CBFB (one each). In addition, 27 gains, 42 losses and 41 copy neutral losses of heterozygosity (CN-LOH) were observed in 58 (37%) patients. Recurrent gains affected regions 6q23.3q23.3 (135.325.751-135.607.060) (n=2) and 8q24.21q24.21 (130.517.732-130.808.381) (n=2) while recurrent losses were found for the regions 21q22.12q22.12 encompassing the RUNX1 gene (36.228.735-36.303.952) (n=5), 5q31.2q31.2 including i.a. EGR1 and CTNNA1 (137.617.569-138.993.959) (n=3), 2q34q34 including i.a. IKZF2 (213.371.237-214.560.488) (n=2), 7q22.1q22.1 encompassing i.a. CUX1 (100.485.221-101.916.623) (n=2), and Yq11.223q12 (24.980.949-28.804.541) (n=2). Recurrent CN-LOH were observed on chromosomes 11q (n=10), 2p (n=5), 4q (n=4), 21q (n=3), 1p (n=2), 17q (n=2) and 18q (n=2). 20/27 gains and 38/42 losses were < 10 megabases in sizes and thus below the resolution of chromosome banding analysis. Only in 7 (4%) patients abnormalities (n=11) were identified which in principle are detectable by chromosome banding analysis. These were missed by chromosome banding analysis due to insufficient in vitro proliferation of the aberrant clone, small clone size or poor chromosome morphology. Comparing age, white blood cell count and bone marrow blast counts revealed no differences between patients with or without abnormalities detected by FISH and/or array CGH. However, FLT3-ITD and WT1 mutations were more frequent in cases with abnormalities (25% vs 7%, p=0.001; 9% vs 1%, p=0.021). Survival analysis was performed for 90 intensively treated patients. Overall survival (OS), event-free survival (EFS) and OS with censoring at time of allogeneic SCT (OSctx) were significantly shorter in patients with abnormalities detected by FISH and/or array CGH compared to those without (median OS: 19 vs 49 months, p=0.027; Figure 1; median EFS: 9 vs 21 months, p=0.016; median OSctx: 13 vs 26 months, p=0.018).
Conclusions: 1. AML with normal karyotype based on chromosome banding analysis lacking a disease defining molecular mutation harbor balanced rearrangements, copy number gains and losses as well as CN-LOH at a high frequency (42%). 2. The presence of these abnormalities has a negative impact on survival demonstrating that FISH and array CGH can add prognostic information in the diagnostic work-up of AML with normal karyotype lacking a disease defining mutation. 3. Further investigations of mutations in genes located in regions of recurrent CN-LOH is necessary.
Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Zieschang:MLL Munich Leukemia Laboratory: Employment. Schnittger:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Alpermann:MLL Munich Leukemia Laboratory: Employment. Zenger:MLL Munich Leukemia Laboratory: Employment. Perglerová:MLL2 s.r.o.: Employment. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal