Abstract
Background and Methods
DLBCL patients exhibit striking heterogeneity in therapeutic responses, yet current methods to assess residual disease have shown suboptimal performance. Circulating tumor DNA (ctDNA) is a promising noninvasive biomarker for disease burden. While High-Throughput Sequencing of immunoglobulin genes (Ig-HTS) has shown utility for ctDNA detection in DLBCL, only a small number of tumor markers are profiled, limiting its sensitivity and broad applicability (Kurtz DM, Blood 2015 and Roschewski M, Lancet Oncology 2015). Here, we studied DLBCL patients using CAPP-Seq, a recently developed approach for ultrasensitive assessment of ctDNA that profiles multiple genomic regions across hundreds of kilobases in a single HTS assay (Newman AM, Nat. Med. 2014).
Results
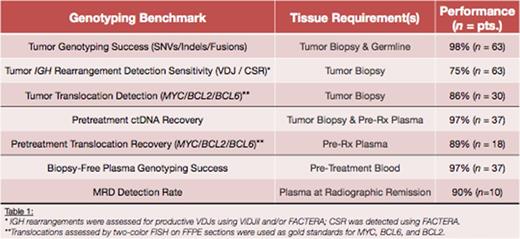
We designed a 250kb DLBCL CAPP-Seq panel targeting 1053 regions from 268 genes, incorporating recurrent single nucleotide variants (SNVs), insertions/deletions (indels) and rearrangements, and evaluated its genotyping performance on diverse tumors from 63 patients (Table 1). We detected somatic mutations in 98% of patients (median: 143 variants/pt), including AID-related events in well-known hotspots. Moreover, we detected 100% of BCL2/BCL6 rearrangements and most MYC translocations previously identified by FISH (n = 30).
Biopsy-confirmed tumor mutations were detectable with 99.3% specificity in 97% of pretreatment plasma samples (n = 37) with a range of 0.007% to 32% mean fractional abundance. Median recovery rates for both SNVs and fusions in plasma exceeded 85%, indicating robust tumor genotyping performance. Furthermore, in most patients, paired allelic fractions between tumor biopsies and pretreatment plasma samples were significantly correlated (P < 0.05), demonstrating that ctDNA can accurately mirror the mutational profiles found in tumor tissues.
We next evaluated the utility of direct biopsy-free genotyping of pretreatment plasma using a novel ctDNA genotyping approach. We noninvasively identified variants in 97% of cases (n=37), and confirmed a median of 95% of SNVs per cfDNA sample in paired tumor biopsies, indicating that plasma DNA is an effective surrogate for direct tumor genotyping.
Having established the technical performance in pretreatment cfDNA, we next assessed utility of CAPP-Seq for minimal residual disease (MRD) detection (Figure 1). Plasma samples at time points of complete response (n = 22) were collected from 10 patients, all of whom ultimately relapsed. Each plasma sample was then analyzed for evidence of biopsy-confirmed mutations from pretreated tumors. Strikingly, ctDNA was detected in 9 of 10 patients (90%), including 3 of 3 isolated CNS relapses, with fractional abundances as low as 0.004%. Among patients with CAPP-Seq detectable MRD, the median time between first detection and relapse was 162 days, and all patients whose MRD levels were uniformly below the detection limit had blood collections >8 months prior to relapse.
Conclusions
To summarize, we validated robust technical performance of a novel approach for noninvasive tumor genotyping in DLBCL using pretreatment plasma, and for MRD surveillance. Given the demonstrated advantages of CAPP-Seq in the setting of radiographic remission, we envision its utility for improved relapse prediction. Moreover, we anticipate that this biopsy-free genotyping approach will have applications for monitoring subclonal dynamics and emergent mutations in response to therapy.
FS, DMK and AMN contributed equally.
Newman:Roche: Consultancy. Klass:Roche: Employment. Diehn:Roche: Consultancy. Alizadeh:Genentech: Consultancy; Celgene: Consultancy; Roche: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal