Key Points
ATF4 positively regulates expansion of functional HSCs in mouse FL.
ATF4-Angptl3 axis in niche cells is pivotal for HSC maintenance in FL.
Abstract
The fetal liver (FL) serves as a predominant site for expansion of functional hematopoietic stem cells (HSCs) during mouse embryogenesis. However, the mechanisms for HSC development in FL remain poorly understood. In this study, we demonstrate that deletion of activating transcription factor 4 (ATF4) significantly impaired hematopoietic development and reduced HSC self-renewal in FL. In contrast, generation of the first HSC population in the aorta-gonad-mesonephros region was not affected. The migration activity of ATF4−/− HSCs was moderately reduced. Interestingly, the HSC-supporting ability of both endothelial and stromal cells in FL was significantly compromised in the absence of ATF4. Gene profiling using RNA-seq revealed downregulated expression of a panel of cytokines in ATF4−/− stromal cells, including angiopoietin-like protein 3 (Angptl3) and vascular endothelial growth factor A (VEGFA). Addition of Angptl3, but not VEGFA, partially rescued the repopulating defect of ATF4−/− HSCs in the culture. Furthermore, chromatin immunoprecipitation assay in conjunction with silencing RNA-mediated silencing and complementary DNA overexpression showed transcriptional control of Angptl3 by ATF4. To summarize, ATF4 plays a pivotal role in functional expansion and repopulating efficiency of HSCs in developing FL, and it acts through upregulating transcription of cytokines such as Angptl3 in the microenvironment.
Introduction
Many efforts have been devoted to investigations of the expansion and maintenance of functional hematopoietic stem cells (HSCs) for therapeutic purposes.1,2 However, to date, none of the developed methods have been firmly demonstrated to be clinically valuable. Thus, deeper understanding of the mechanisms by which HSCs are generated, amplified, and maintained in developing embryos may guide the future development of more effective techniques for therapeutic manipulations of HSCs.
The process of embryonic hematopoiesis can be separated into 2 stages: primitive and definitive hematopoiesis. Definitive hematopoiesis is characterized by the generation of adult-type HSCs in midgestation mouse embryos. HSCs form in the aorta-gonad-mesonephros (AGM) region at embryonic day 10.5 (E10.5) and subsequently migrate into the fetal liver (FL) at E11.5.3,4 At E15.5, these HSCs are released into the circulating blood and begin to home to the bone marrow.5 Of note, the number of HSCs increases drastically from 2 to 3 to 800 to 1000 in mouse FL,4 suggesting a unique and powerful effect of the FL microenvironment on the expansion of HSCs. Therefore, identifying novel regulators and microenvironment cues for HSC development in FL is of great importance.
In mouse FL, stromal cells, hepatoblasts, and endothelial cells are the 3 main cell types that significantly contribute to the extrinsic regulation of HSC development.6 Among them, stromal cells harvested from either primary culture or immortalized cell lines express both mesenchymal markers (eg, vimentin, osteopontin, and alpha smooth muscle actin) and epithelial markers (eg, α-fetoprotein, cytokeratins 8, and albumin) and are able to support hematopoiesis.7 In addition, the stem cell factor (SCF)+DLK+ hepatoblasts maintain the activity of HSCs by producing cytokines such as thrombopoietin (TPO), SCF, angiopoietin-like protein 3 (Angptl3), and insulin-like growth factor 2 (IGF2).8 Moreover, endothelial cells secrete the chemokine CXCL12 to promote HSC maintenance.9 However, the core regulatory mechanism, particularly the transcriptional program that operates in mosaic niche cells and is crucial for HSC development in FL is largely unknown.
Activating transcription factor 4 (ATF4) is a basic region-leucine zipper transcription factor that is widely expressed in many tissues and cells and functions as a stress response factor and a developmental regulator.10 The absence of ATF4 induces partial perinatal lethality11 and results in severe anemia and abnormal erythropoiesis in E15.5 embryos.12,13 However, whether ATF4 plays a critical role in definitive hematopoiesis at the stem cell level, such as de novo generation, migration, amplification, and maintenance of HSCs, is a logical and important question to be answered.
In this study, we show that ATF4 deletion does not affect initial HSC generation in the AGM region but markedly impairs the expansion of functional HSCs in the FL. Mechanistically, ATF4 can transcriptionally upregulate expression of Angptl3 in the niche cells, which may guarantee expansion and maintenance of functional HSCs during the unique wave of FL hematopoiesis.
Methods
Mice
B6-Ly5.2 and B6-Ly5.1 mice were purchased from the animal facility of the State Key Laboratory of Experimental Hematology. The ATF4+/− mice were obtained from Dr Guozhi Xiao.14 We backcrossed the ATF4+/− mice to B6-Ly5.2 mice 10 times and obtained ATF4 knockout (KO; ATF4−/−) and wild-type (WT; ATF4+/+) control embryos using ATF4+/− mice (CD45.2). The experimental protocol was approved by the Institutional Animal Care and Use Committees of State Key Laboratory of Experimental Hematology. To genotype the mice, DNA was extracted from the tail tips; detailed procedures are presented in the supplemental Data available on the Blood Web site.
Embryo dissection and single-cell isolation
Flow cytometry
FL cells were incubated in various antibodies for 30 minutes in the dark at 4°C. Stained cells were analyzed via fluorescence-activated cell sorter (FACS) by using a FACSCalibur flow cytometer or were sorted using a FACSAria II flow cytometer (BD Biosciences).
cBMT
The competitive bone marrow transplantation (cBMT) experiments were performed as previously described.17 More details can be found in the supplemental Data.
Accession number
The microarray and RNA-seq data are available in the Gene Expression Omnibus database under the accession number GSE66382.
Statistical analysis
The significance of differences was assessed by using Student t test. The data are presented as the means ± standard error of the mean (SEM).
Additional experimental procedures are described in the supplemental Data.
Results
ATF4 deletion severely impaired the hematopoietic repopulating potential of FL cells
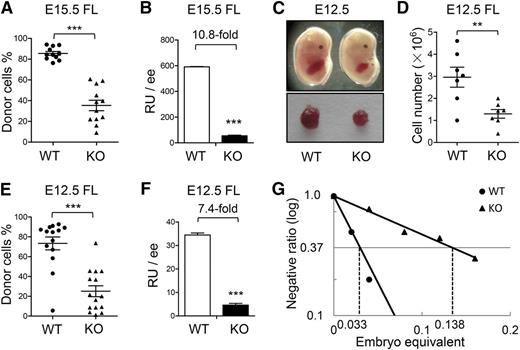
It was previously reported that the E15.5 ATF4−/− FL had lower numbers of total nucleated cells and colony-forming cells (CFCs) than the ATF4+/+ control.12 To further examine in vivo hematopoietic repopulating potential, we performed the cBMT assay by transplanting 0.01 embryo equivalent (ee) of E15.5 ATF4+/+ or ATF4−/− FL cells (CD45.2) together with 105 bone marrow nucleated cells (BMNCs) from the congenic mice (CD45.1) into lethally irradiated recipients (CD45.1). At 16 weeks after transplantation, the donor chimerism in recipient peripheral blood of the ATF4−/− group was 35.3% ± 5.0%, significantly lower than that of the ATF4+/+ counterpart (85.5% ± 1.8%) (WT: n = 11; KO: n = 12; 3 independent experiments) (Figure 1A). On the basis of the chimerism, we calculated the repopulating units (RUs).17,18 Assuming that 105 BMNCs is one RU, we found a 10.8-fold reduction in the ATF4−/− group (Figure 1B). However, the lineage differentiation spectrum of the ATF4−/− donor cells was not significantly affected in peripheral blood and bone marrow of recipients (supplemental Figure 1A-B).
Impaired HSC development in the ATF4−/− FL. (A-B) Donor chimerism and RUs of E15.5 FL cells at 16 weeks after transplantation (n ≥ 11 for each genotype; 3 independent experiments; WT: ATF4+/+; KO: ATF4−/−). (C) Photographs of the embryos and FLs at E12.5. (D) Cell number per FL at E12.5 (n = 7 for each group). (E-F) Donor chimerism and RUs of E12.5 FL cells at 16 weeks after transplantation (n ≥ 14 for each group; 3 independent experiments). (G) Limiting dilution analysis of reconstitution ability in E12.5 FL. The following dose gradients were used: 0.02, 0.04, 0.08, 0.12, and 0.16 ee (n ≥ 9 for each group; 2 independent experiments). Student t test **P < .01 and ***P < .001.
Impaired HSC development in the ATF4−/− FL. (A-B) Donor chimerism and RUs of E15.5 FL cells at 16 weeks after transplantation (n ≥ 11 for each genotype; 3 independent experiments; WT: ATF4+/+; KO: ATF4−/−). (C) Photographs of the embryos and FLs at E12.5. (D) Cell number per FL at E12.5 (n = 7 for each group). (E-F) Donor chimerism and RUs of E12.5 FL cells at 16 weeks after transplantation (n ≥ 14 for each group; 3 independent experiments). (G) Limiting dilution analysis of reconstitution ability in E12.5 FL. The following dose gradients were used: 0.02, 0.04, 0.08, 0.12, and 0.16 ee (n ≥ 9 for each group; 2 independent experiments). Student t test **P < .01 and ***P < .001.
Because HSCs undergo drastic expansion in normal mouse FL at approximately E12.5,4 we asked whether ATF4 deficiency had a negative impact on hematopoiesis at this time point. As shown in Figure 1C-D, the E12.5 ATF4−/− fetuses appeared anemic and contained smaller and paler FLs with lower cellularity than the ATF4+/+ control (1.3 ± 0.2 × 106 vs 3.0 ± 0.5 × 106). Next, we separately transplanted 0.08 ee of the 2 genotypes per recipient and observed a reduced reconstitution ability of E12.5 ATF4−/− FL cells (KO: 25.1% ± 5.5% vs WT: 73.4% ± 6.5%), indicating a 7.4-fold decrease in RUs (Figure 1E-F). As observed at E15.5, the multilineage differentiation potential of ATF4−/− FL cells was unaltered in E12.5 embryos.
To enumerate the absolute number of functional HSCs, competitive RU (CRU) was calculated with a limiting dilution assay using E12.5 FL cells. On the basis of 16-week posttransplant readouts with varied numbers of input of ee, we found that there were 7.25 and 30.30 CRUs per ATF4−/− or ATF4+/+ FL, respectively (Figure 1G), indicating a 4.2-fold decrease of functional HSCs in the ATF4−/− FL at E12.5.
Based on the numbers of RUs and CRUs, we calculated the mean activity per stem cell, which is equal to RU/CRU.19 There was an approximately 1.8-fold reduction in repopulating potential of each HSC from E12.5 ATF4−/− FL (supplemental Table 1), suggesting a decreased yield of hematopoietic progenitor cells (HPCs). The CFC assay consistently demonstrated that the size of ATF4−/− colonies was markedly decreased (supplemental Figure 2A). In addition, in the ATF4−/− group, we observed a sharply decreased number of granulocyte, erythrocyte, monocyte, and macrophage CFCs (KO: 550.0 ± 75.4 vs WT: 1133.0 ± 95.5 at day 7) and high proliferative potential colonies (KO: 366.7 ± 33.3 vs WT: 666.7 ± 88.2 at day 14) (supplemental Figure 2B), indicating that ATF4 deletion could also significantly perturb the function of early HPCs in the E12.5 FL.
Therefore, in addition to HPCs, development of HSCs was severely impaired in the ATF4−/− FL, as witnessed by 7.4-fold to 10.8-fold reduction in RUs from E12.5 to E15.5. This was largely a result of the paucity of functional HSCs (4.2-fold reduction in CRUs at E12.5) and was also caused by compromised repopulation per HSC (1.8-fold reduction in mean activity per stem cell).
ATF4 deletion did not affect HSC generation in the AGM region
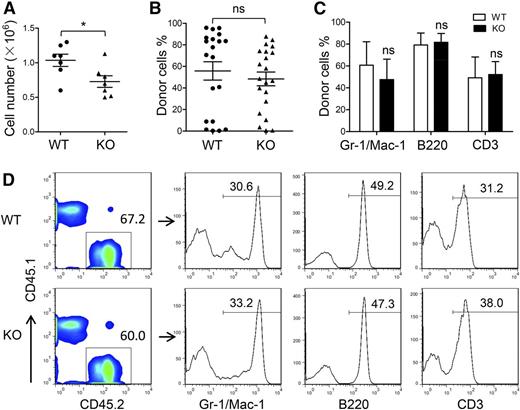
The AGM region is considered the predominant site of de novo generation of HSCs in mice. We determined whether the HSC defect in ATF4−/− FL was caused by insufficient generation of HSCs in the AGM region. At E11.5, the ATF4−/− AGM region contained slightly fewer cells than the ATF4+/+ AGM region (Figure 2A). Despite this fact, when we injected single-cell suspensions consisting of 1 ee of the AGM region into each recipient, we found that the mean chimerism was not significantly different between the 2 groups (WT: 55.8% ± 6.4%, n = 20; KO: 48.4% ± 8.6%, n = 21; 6 independent experiments) (Figure 2B). In addition, the ATF4−/− donor cells could differentiate into myeloid cells, B lymphocytes, and T lymphocytes in the peripheral blood, bone marrow, spleen, and thymus in the recipient mice (Figure 2C-D and supplemental Figure 3A-F).
Uninfluenced generation of HSCs in the ATF4−/− E11.5 AGM region. (A) Cell number per AGM region at E11.5 (n = 7 for each group). (B) Donor chimerism of E11.5 AGM region at 24 weeks after transplantation from 6 independent experiments (n ≥ 20 for each group). (C) Donor-derived multilineage chimerism in the peripheral blood of recipients at 24 weeks after transplantation (n ≥ 5 for each group). (D) Composition of myeloid (Mac-1 and/or Gr-1), B lymphoid (B220), and T lymphoid (CD3) donor cells (CD45.2) in peripheral blood at 24 weeks after transplantation. Student t test *P < .05 and **P < .01. ns, not significant.
Uninfluenced generation of HSCs in the ATF4−/− E11.5 AGM region. (A) Cell number per AGM region at E11.5 (n = 7 for each group). (B) Donor chimerism of E11.5 AGM region at 24 weeks after transplantation from 6 independent experiments (n ≥ 20 for each group). (C) Donor-derived multilineage chimerism in the peripheral blood of recipients at 24 weeks after transplantation (n ≥ 5 for each group). (D) Composition of myeloid (Mac-1 and/or Gr-1), B lymphoid (B220), and T lymphoid (CD3) donor cells (CD45.2) in peripheral blood at 24 weeks after transplantation. Student t test *P < .05 and **P < .01. ns, not significant.
These data demonstrated that the hematopoietic ability of the E11.5 AGM region was not influenced by ATF4 deletion, suggesting that ATF4 deficiency did not affect the formation of the first population of HSCs prior to their colonization in the FL. The slightly lower cellularity in the ATF4−/− AGM region might be attributed to the nonhematopoietic cells. Therefore, the decreased number and reduced function of HSCs in the FL in the absence of ATF4 were unlikely the result of a change in HSC genesis from the AGM region.
HSC migration was moderately suppressed in the absence of ATF4
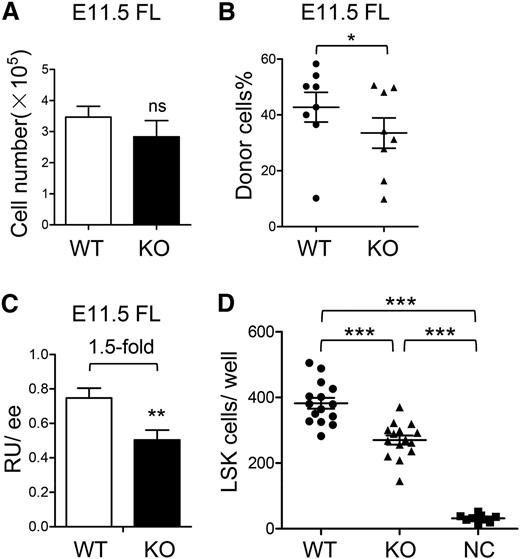
Upon emergence of HSCs in the AGM region, they migrate into circulating blood and colonize the FL. Because the functional defect of HSCs generated in the AGM region was not observed, we then asked whether ATF4 deletion might negatively regulate the entry of HSCs into FL. To this end, we analyzed the FL at E11.5, the earliest time point when HSCs begin to reside there and can be detected.4 The ATF4−/− FL did not show significant reduction in cellularity at E11.5 as opposed to later time points (Figure 3A). When 1 ee of E11.5 FL cells (CD45.2) was transplanted to the recipients with 1 × 105 competitor cells (CD45.1) in the cBMT assay, there was a 1.5-fold reduction of RUs in the ATF4−/− group (Figure 3B-C), suggesting the initial number of functional HSCs had a moderate decrease in ATF4−/− FL.
Moderately suppressed migration of HSCs in the absence of ATF4. (A) Cell number per FL at E11.5 (n = 4 for each group). (B-C) Donor chimerism and RU of E11.5 FL cells at 12 weeks after transplantation (n = 8 for each group). (D) Transwell assay of 104 LSK cells in the ATF4+/+ or ATF4−/− FL. Cell counts in the lower chamber are shown (n ≥ 9 for each group; 3 independent experiments). Student t test *P < .05, **P < .01, and ***P < .001.
Moderately suppressed migration of HSCs in the absence of ATF4. (A) Cell number per FL at E11.5 (n = 4 for each group). (B-C) Donor chimerism and RU of E11.5 FL cells at 12 weeks after transplantation (n = 8 for each group). (D) Transwell assay of 104 LSK cells in the ATF4+/+ or ATF4−/− FL. Cell counts in the lower chamber are shown (n ≥ 9 for each group; 3 independent experiments). Student t test *P < .05, **P < .01, and ***P < .001.
To further examine the migration capacity of immunophenotypically defined HSCs in vitro, the transwell assay was performed; 1 × 104 ATF4+/+ or ATF4−/− Lin–Sca-1+c-Kit+ (LSK) cells at E15.5 were deposited onto each upper chamber of transwell inserts, and lower chambers were filled with nutritious medium. Four hours later, we calculated cell number in each lower chamber and found a 1.4-fold reduction in the ATF4−/− group compared with the ATF4+/+ group (270.4 ± 14.0 vs 382.3 ± 16.4) (Figure 3D). To a certain extent, the moderate reduction in HSC migration might contribute to defective development of HSCs in ATF4−/− FL.
ATF4 deletion dampened the self-renewal potential of HSCs from FL
The moderate decrease in HSC migration does not appear to play a major role in the impairment of hematopoiesis in FL in the absence of ATF4. Instead, the paucity of functional HSCs in ATF4−/− FL was likely the result of defective self-renewal capacity of HSCs. To address this issue, serial transplantation starting with the same input number of purified HSCs was conducted.
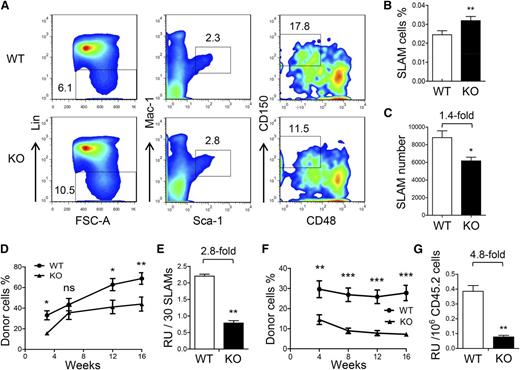
At first, we analyzed the frequency and number of HSCs in FL at different time points according to previous studies. We determined the HSC frequency by analyzing the CD201+LSK cells at E12.5, Lin–Sca-1+Mac-1lowCD150+CD48– cells at E14.5, and CD48–CD150+LSK cells at E15.5.9,20,21 At the three time points analyzed, the HSC frequencies were all slightly increased (Figure 4A-B and supplemental Figure 4A-D), but the numbers were reduced by approximately 1.5-fold in the ATF4−/− group (Figure 4C and supplemental Figure 4E-F).
Decreased self-renewal ability of HSCs in the ATF4−/− FL. (A) Representative flow cytometric analysis of HSCs in the FL at E14.5. FSC-A, forward-scattering angle. (B-C) Frequency and number of SLAM cells in the FL at E14.5 (n = 4 for each genotype). (D-E) Donor chimerism and RU of 30 SLAM cells at different time points after transplantation (n = 12 for each group). (F) Donor chimerism of secondary transplantation using 1 × 106 donor-derived BMNCs (CD45.2) and 1 × 105 competitor BMNCs (CD45.1). (G) Number of RUs in 1 × 106 primary donor cells at 16 weeks after secondary transplantation (n = 12). Student t test *P < .05, **P < .01, and ***P < .001.
Decreased self-renewal ability of HSCs in the ATF4−/− FL. (A) Representative flow cytometric analysis of HSCs in the FL at E14.5. FSC-A, forward-scattering angle. (B-C) Frequency and number of SLAM cells in the FL at E14.5 (n = 4 for each genotype). (D-E) Donor chimerism and RU of 30 SLAM cells at different time points after transplantation (n = 12 for each group). (F) Donor chimerism of secondary transplantation using 1 × 106 donor-derived BMNCs (CD45.2) and 1 × 105 competitor BMNCs (CD45.1). (G) Number of RUs in 1 × 106 primary donor cells at 16 weeks after secondary transplantation (n = 12). Student t test *P < .05, **P < .01, and ***P < .001.
In this study, we relied more on functional assessment than phenotypic quantitation of HSCs. Therefore, we sorted the immunophenotypically defined HSC population in the E14.5 ATF4−/− FL by using the signaling lymphocyte activation molecule (SLAM) markers for mouse HSCs21 and then transplanted 30 SLAM cells (CD45.2) along with competitor cells (CD45.1) into each recipient (CD45.1). After 16 weeks, we found that the long-term reconstituting ability of the ATF4−/− SLAM cells was significantly decreased by 2.8-fold (Figure 4D-E). Furthermore, after 16 weeks, secondary transplantation was performed by using 1 × 106 donor BMNCs (CD45.2) sorted from repopulated primary recipients with 1 × 105 BMNCs (CD45.1) as competitors. Of note, in the primary recipients, the percentage of SLAM cells in the BMNCs (CD45.2) was slightly lower in the ATF4−/− group (KO: 2.0 ± 0.2 × 10−4 vs WT: 2.6 ± 0.2 × 10−4) (supplemental Figure 5A-B). At 16 weeks after secondary transplantation, the RU of donor cells was further decreased by 4.8-fold in the ATF4−/− group (KO: 0.08 ± 0.01 vs WT: 0.38 ± 0.04), indicating a reduced self-renewal potential of the ATF4−/− HSCs (Figure 4F-G). This was highly consistent with the result obtained from a separate serial transplantation using the same input number of bulk cell populations (unpurifed HSCs) (supplemental Figure 6A-C).
To exclude the possibility that the lower reconstitution rate might be the result of compromised homing ability in the transplant recipients, we sorted 1 × 105 HSC-enriched LSK cells and transplanted them into each lethally irradiated recipient (CD45.1). After 17 hours, BMNCs were analyzed by FACS. The abundance of homed LSK cells (CD45.2) was calculated, and no significant difference could be found between the 2 genotypes (supplemental Figure 7A-B).
The ability of microenvironmental cells to support HSCs in the ATF4−/− FL was significantly decreased
To investigate the molecular basis for the functional defects of ATF4−/− HSCs, we isolated the LSK cells from ATF4−/− or ATF4+/+ FL (from 4 independent experiments) to analyze their global gene expression profiles via microarray. The Kyoto Encyclopedia of Genes and Genomes pathway analysis of the differentially expressed genes revealed the enrichment of 10 pathways, 3 of which were related to molecules in the FL microenvironment: cell adhesion molecules (P = 4.96e-13), cytokine-cytokine receptor interaction (P = 9.94e-10), and extracellular matrix receptor interaction (P = 3.38e-6) (supplemental Table 2). Therefore, these data encouraged us to focus more on microenvironmental effects on HSCs in FL.
Endothelial cells and stromal cells constitute the primary components of the FL niche for hematopoietic cells.6 Thus, we isolated the endothelial (CD31+CD45–Ter119–) or stromal (CD31–CD45–Ter119–) cells from ATF4−/− and ATF4+/+ FL and analyzed the apoptosis, cell cycle status, and senescence in these cells. No overt differences were observed in the endothelial cells (supplemental Figure 8A-C,I), but a moderate increase in the apoptosis of ATF4−/− stromal cells was detected (KO: 7.7% ± 1.6% vs WT: 3.6% ± 0.8%) (supplemental Figure 8D-I). To determine the HSC-supporting ability of the 2 different cell subsets, we performed the assay by coculturing wild-type ATF4+/+ SLAM cells with these cells (Figure 5A-B). After 3 to 4 days, the cell number was similar but the LSK frequency was higher in the ATF4+/+ coculture than in the ATF4−/− counterpart (Figure 5C-F). We subsequently transplanted these cultured cells into lethally irradiated recipients (CD45.1). At 16 weeks after transplantation, we found that the chimera ratio was significantly decreased in ATF4−/− endothelial cell coculture (30.3% ± 3.6%) compared with the ATF4+/+ control (61.0% ± 8.4%) (Figure 5G). A similar result was found in the stromal cell coculture (KO: 7.8% ± 2.4%; WT: 20.8% ± 4.7%) (Figure 5H). Thus, the HSC-supporting ability of the ATF4−/− FL microenvironment was significantly reduced.
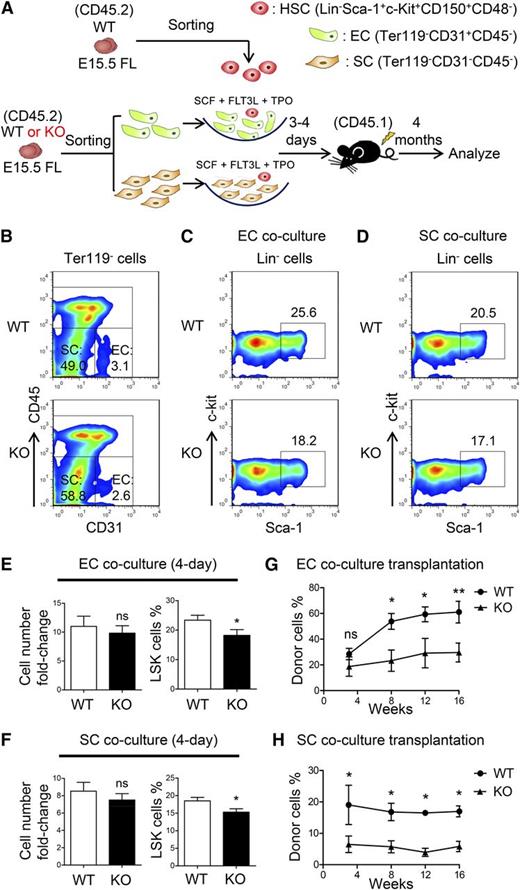
Decreased HSC-supporting ability of niche cells in the ATF4−/− FL. (A) Schematic illustration of the experimental designused to determine the effect of endothelial cells (ECs) or stromal cells (SCs) on WT HSCs (SLAM cells). (B) Representative flow cytometric analysis of the endothelial and stromal cells detected in the FL. (C-F) Representative flow cytometric analysis and statistical data of WT HSCs cocultured with endothelial or stromal cells for 3 to 4 days (n = 3 for each group). (G-H) Donor chimerism of the WT HSCs cocultured with endothelial or stromal cells of different genotypes for 3 to 4 days at 16 weeks after transplantation (n ≥ 7). Student t test *P < .05 and **P < .01.
Decreased HSC-supporting ability of niche cells in the ATF4−/− FL. (A) Schematic illustration of the experimental designused to determine the effect of endothelial cells (ECs) or stromal cells (SCs) on WT HSCs (SLAM cells). (B) Representative flow cytometric analysis of the endothelial and stromal cells detected in the FL. (C-F) Representative flow cytometric analysis and statistical data of WT HSCs cocultured with endothelial or stromal cells for 3 to 4 days (n = 3 for each group). (G-H) Donor chimerism of the WT HSCs cocultured with endothelial or stromal cells of different genotypes for 3 to 4 days at 16 weeks after transplantation (n ≥ 7). Student t test *P < .05 and **P < .01.
RNA-seq of ATF4−/− stromal cells showed decreased expression of Angptl3 and VEGFA
To further elucidate the molecular basis by which the supporting ability of these stromal cells was defective, RNA-seq was performed on these cells harvested from 5 independent experiments, and the gene ontology terms of the gene set were analyzed by using an established cytoscape plug-in: Enrichment Map.22 The downregulated genes were functionally enriched in 4 main biological processes: lipid process, inflammatory response, response to stimulus, and coagulation (Figure 6A).
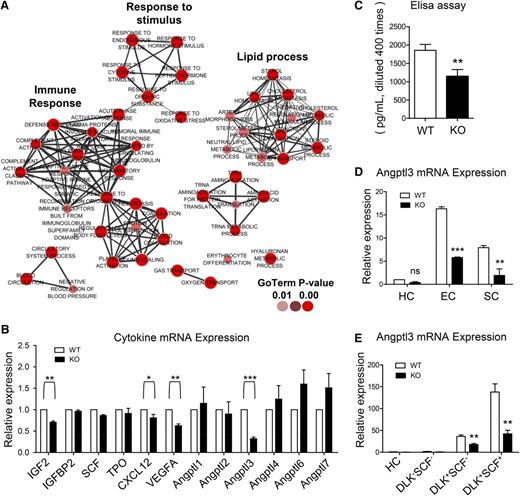
Drastically decreased expression of Angptl3 in the ATF4−/− FL. (A) Gene ontology (GO) terms corresponding to the downregulated genes in the stromal cells (harvested from 5 independent experiments) based on RNA-seq. (B) Expression of the genes related to HSC expansion and Angptl family genes in the E15.5 FL stromal cells from 3 independent experiments. (C) Expression of Angptl3 protein in the E15.5 FL homogenates based on enzyme-linked immunosorbent assay (ELISA) from 3 independent experiments. (D) Expression of Angptl3 mRNA in hematopoietic cells (HCs), ECs, and SCs by real-time PCR from 3 independent experiments. (E) Expression of Angptl3 mRNA in HCs, DLK–SCF– cells, DLK+SCF– cells, and DLK+SCF+ cells (hepatoblasts) by real-time PCR from 3 independent experiments. Student t test *P < .05, **P < .01, and ***P < .001.
Drastically decreased expression of Angptl3 in the ATF4−/− FL. (A) Gene ontology (GO) terms corresponding to the downregulated genes in the stromal cells (harvested from 5 independent experiments) based on RNA-seq. (B) Expression of the genes related to HSC expansion and Angptl family genes in the E15.5 FL stromal cells from 3 independent experiments. (C) Expression of Angptl3 protein in the E15.5 FL homogenates based on enzyme-linked immunosorbent assay (ELISA) from 3 independent experiments. (D) Expression of Angptl3 mRNA in hematopoietic cells (HCs), ECs, and SCs by real-time PCR from 3 independent experiments. (E) Expression of Angptl3 mRNA in HCs, DLK–SCF– cells, DLK+SCF– cells, and DLK+SCF+ cells (hepatoblasts) by real-time PCR from 3 independent experiments. Student t test *P < .05, **P < .01, and ***P < .001.
Some of the downregulated genes such as Angptl3, vascular endothelial growth factor A (VEGFA), and IGF2 were previously demonstrated to be involved in hematopoiesis.1,23,24 Thus, the expression of these genes and some other known hematopoietic cytokines (SCF, TPO, IGFBP2, CXCL12, and other Angptl family members) in FL hematopoiesis was verified by real-time polymerase chain reaction (PCR).25-27 Notably, the expression levels of Angptl3 and VEGFA were significantly decreased by 68% and 38%, respectively, in the ATF4−/− group (Figure 6B), implying that ATF4 has a role in the transcriptional regulation of these cytokines.
Angptl3 has been reported to support the activity of HSCs in bone marrow and FL and to be expressed by HSC-supportive fetal hepatoblasts.8,27,28 As shown in Figure 6C, the protein levels of Angptl3 in the ATF4−/− FL homogenates were decreased. To determine the cellular source of Angptl3, we isolated hematopoietic (CD45+Ter119–), endothelial (CD31+CD45–Ter119–), and stromal (CD31–CD45–Ter119–) cells via flow cytometry. The endothelial cells and the stromal cells displayed abundant expression of Angptl3, and in contrast, the hematopoietic cells displayed scarce expression of Angptl3 (Figure 6D). Interestingly, the expression of Angptl3 was clearly reduced in the ATF4−/− FL compared with the ATF4+/+ FL in both endothelial cells and stromal cells.
Because hepatoblasts could secret Angptl3 abundantly,29 we analyzed them in the FL. Interestingly, we found that hepatoblasts mainly existed in the Ter119–CD45–CD31– stromal cell population, and their frequencies were similar between the 2 genotypes (supplemental Figure 9A). Real-time PCR analysis showed that the expression of Angptl3 messenger RNA (mRNA) was decreased in the ATF4−/− DLK+SCF+ hepatoblasts, similar to that in stromal cells (Figure 6E). Therefore, we suspected that the ATF4−/− hepatoblasts should be an important source of the Angptl3 that was responsible for impaired function of HSCs in the aberrant FL microenvironment.
Angptl3, but not VEGFA, partially rescued the defects of ATF4−/− HSCs
To further examine whether Angptl3 protein enhances the function of HSCs in the absence of ATF4, SLAM cells were sorted from the ATF4−/− FL and cultured with SCF, FLT3L, and TPO in the presence or absence of Angptl3. After 4 days in culture, although the total cell numbers were similar, the frequencies of LSK cells (20.2% ± 2.5% vs 18.5% ± 2.9%) and CD48–LSK cells (12.1% ± 0.7% vs 9.4% ± 0.8%) were higher in the Angptl3-treated group than in the group that lacked Angptl3 (Figure 7A-B). Then we separately collected all of the cultured cells and injected these cells into lethally irradiated recipients (9 recipients per group from 3 independent experiments). At 16 weeks after transplantation, we found that the reconstitution ratio (chimerism >10%) was higher in the Angptl3-treated group than in the control group (6 of 9 vs 3 of 9) (Figure 7C). In contrast, no significant differences in the LSK frequency after 4 days of coculture or in reconstitution ability at 16 weeks after transplantation were observed between the VEGFA-treated group and the group that lacked VEGFA (supplemental Figure 10A-B), suggesting that VEGFA could not rescue the defective function of the ATF4−/− HSCs.
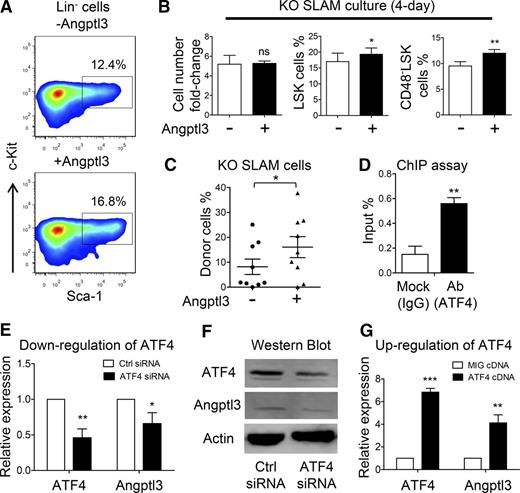
Restorative effect of Angptl3 on functional deficiency of HSCs from the ATF4−/− FL. (A) Representative flow cytometric analysis of LSK cells cultured in the presence or absence of Angptl3 for 4 days (3 independent experiments). (B) Fold change of cell number and percentage of LSK and CD48–LSK cells after culturing ATF4−/− HSCs with or without Angptl3 for 4 days (3 independent experiments). (C) Donor chimerism of the ATF4−/− SLAM cells cultured with or without Angptl3 for 4 days at 16 weeks after transplantation (3 independent experiments). (D) Percentage of input of the ATF4 antibody using mouse FL cells based on the chromatin immunoprecipitation (ChIP) assay. Primers were designed to amplify the Angptl3 promoter (n = 3 for each group). Expression of (E) Angptl3 mRNA or (F) protein by transfecting ATF4 siRNA into an NIH/3T3 cell line (3 independent experiments). (G) Expression of Angptl3 mRNA by transfecting ATF4 complementary DNA (cDNA) overexpression vector into an NIH/3T3 cell line (4 independent experiments). Student t test *P < .05, **P < .01, and ***P < .001. Ab, antibody; IgG, immunoglobulin G; MIG, mouse stem cell virus-internal ribosome entry site-green fluorescent protein.
Restorative effect of Angptl3 on functional deficiency of HSCs from the ATF4−/− FL. (A) Representative flow cytometric analysis of LSK cells cultured in the presence or absence of Angptl3 for 4 days (3 independent experiments). (B) Fold change of cell number and percentage of LSK and CD48–LSK cells after culturing ATF4−/− HSCs with or without Angptl3 for 4 days (3 independent experiments). (C) Donor chimerism of the ATF4−/− SLAM cells cultured with or without Angptl3 for 4 days at 16 weeks after transplantation (3 independent experiments). (D) Percentage of input of the ATF4 antibody using mouse FL cells based on the chromatin immunoprecipitation (ChIP) assay. Primers were designed to amplify the Angptl3 promoter (n = 3 for each group). Expression of (E) Angptl3 mRNA or (F) protein by transfecting ATF4 siRNA into an NIH/3T3 cell line (3 independent experiments). (G) Expression of Angptl3 mRNA by transfecting ATF4 complementary DNA (cDNA) overexpression vector into an NIH/3T3 cell line (4 independent experiments). Student t test *P < .05, **P < .01, and ***P < .001. Ab, antibody; IgG, immunoglobulin G; MIG, mouse stem cell virus-internal ribosome entry site-green fluorescent protein.
In light of a previous study showing that the expression of Angptl is regulated by ATF family members,30,31 we next examined whether ATF4 exerts this effect on Angptl3. We first performed a chromatin immunoprecipitation (ChIP) assay and found that ATF4 directly binds to the Angptl3 promoter (Figure 7D). Next, we silenced ATF4 expression in the NIH/3T3 cell line by using ATF4 silencing RNA (siRNA), and the real-time PCR results showed that the mRNA levels of Angptl3 were decreased as a result of treatment with ATF4 siRNA compared with control siRNA (Figure 7E). Furthermore, we performed a western blot analysis and found that the protein level of Angptl3 was also reduced by ATF4 siRNA (Figure 7F). The overexpression of ATF4 complementary DNA consistently elevated the expression of Angptl3 mRNA (Figure 7G). Taken together, our results suggested that ATF4 may play a direct role in the transcriptional regulation of Angptl3 in FL cells.
Discussion
It is well known that the FL serves as an important and unique site for rapid amplification of functional HSCs during development, but its underlying mechanism has been poorly understood. In this study, we found that ATF4 is a potent positive regulator for the functional expansion and repopulating efficiency of HSCs in mouse developing FL. In contrast, it does not appear to significantly affect HSC genesis in the AGM region and exerts a moderate effect on HSC entry into FL.
In adult bone marrow, the HSC niche contains the osteoblast-abundant endosteal region in junction with the nearby endothelial vascular region.32 ATF4 has been shown to promote osteoblastic development33 as well as vascularization in bone marrow.14 Thus, because these known niche components are altered in the absence of ATF4, ATF4 is likely required for maintaining the HSC-supportive niche throughout ontogeny and adulthood, despite presumably different molecular circuits. In this study, the expression of some cytokines, including Angptl3, VEGFA, and IGF2, which are known to be involved in HSC development or regulation, was decreased in ATF4−/− FL microenvironmental cells.1,23,28,29 Moreover, Angptl3 was able to particularly rescue the defective functions of ATF4−/− HSCs in the coculture system. Previously, Angptl3 was consistently reported to maintain HSC stemness.1 Thus, ATF4 in hematopoietic supportive cells might ensure the HSC self-renewal property by increasing the expression of Angptl3. However, adding VEGFA to the coculture did not have a restorative effect on ATF4−/− HSCs despite the fact that single-allele deletion of VEGFA was shown to induce the lethality of heterozygous embryos because of severe defects in vasculogenesis and angiogenesis.34 Therefore, VEGFA may play an indirect role, if it plays any role at all, in HSC maintenance by decreasing the number and function of endothelial cells in the developing FL.14
Apart from the microenvironment, HSCs in the ATF4−/− FL may also have intrinsic defects. HSC transplantation data have clearly shown that the reconstitution capacity of ATF4−/− HSCs was significantly reduced. There are at least 2 reasons for this outcome. First, Our data on CRUs clearly demonstrated that HSC self-renewal capacity was decreased as a result of ATF4 deletion. However, phenotypic quantitation did not correlate well with the CRU data. This discrepancy may be the result of either imperfection of the SLAM markers for FL HSCs (similar to that for bone marrow HSCs) or the ongoing development of functional HSCs in FL. Second, there is a defect in the differentiation potential of HSCs, meaning decreased function per HSC. Interestingly, the ATF4−/− HSCs showed the same balanced lineage distribution as the WT HSCs after bulk BMNC or HSC transplantation. Because the donor-derived chimerism was significantly lower in the ATF4−/− group, we speculate that the mutant HSCs may have a defect at an early rather than a late stage of the differentiation cascade, which would result in fewer multipotent progenitors possessing normal differentiation potential to downstream lymphomyeloid lineages.
Notably, given our data showing no significant changes in apoptosis, cell cycle, or senescence of the HSCs in the absence of ATF4 (supplemental Figure 11A-F), one possibility would be that, in the absence of ATF4, the mutant HSCs disfavor symmetric self-renewing division during the amplification of HSCs in FL as a result of both intrinsic and extrinsic defects. In addition, our microarray data also found that Runt-related transcription factor 1 translocated to 1 (Runx1t1) was significantly decreased in ATF4−/− LSK cells. In addition to its classical essential role in definitive hematopoiesis, Runx1t1 was recently shown to be present in a cocktail of several transcription factors that was able to reprogram murine HPCs into functional HSCs.35 In addition, the unfolded protein response has recently been shown to determine the integrity of the HSC pool during endoplasmic reticulum stress during which the expression of ATF4 in HSCs was higher than that in HPCs.36 Therefore, whether ATF4 might regulate the HSC pool partially via unfolded protein response will be of interest in future studies. These intriguing possibilities all deserve further investigation.
Migration is critical for the dynamic development of HSCs in mouse embryos. Interestingly, we found that HSC migration was moderately affected by ATF4 deletion. In support of that notion, gene profiling of HSCs has demonstrated perturbed expression of many genes related to migration, and ATF4 has been reported to stimulate migration of breast cancer cells and promote metastasis of esophageal squamous cell carcinoma.37,38
In summary, this study reveals a new paradigm in which ATF4, as a master transcription factor, determines HSC function by controlling the expression of Angptl3 and likely some other soluble factors in the mosaic niche cells. Therefore, these results have important implications for the development of new strategies for therapeutic HSC expansion.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the laboratories of H.C. and D.L. for their contributions to this study and Professors Chengcheng Zhang and Junke Zheng for their valuable input.
This work was supported by grants 2011CB964800, 2012CB966904, and 2013CB966902 from the Ministry of Science and Technology of China and by grants 81090411, 81421002, 31425012, 81330015, 31371185, 81328004, 81300373, and 81400076 from the National Natural Science Foundation of China.
Authorship
Contribution: T.C. conceived the study; Y.Z., B.L., and T.C. designed the study; Y.Z., J.Z., F.D., X.M., W.W., Y.N., Z.L., Y.W., and Y.P. performed the experiments; Y.Z., D.L., H.C., H.X., Q.-F.W., G.X., and S.M. analyzed the data; and Y.Z., H.C., S.H., X.W., W.Y., B.L., and T.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tao Cheng, Peking Union Medical College, State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, 288 Nanjing Rd, Tianjin 300020, China; e-mail: chengtao@ihcams.ac.cn; and Bing Liu, Translational Medicine Center of Stem Cells, Affiliated Hospital, Academy of Military Medical Sciences, Beijing 100071, China; e-mail: bingliu17@yahoo.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal