In this issue of Blood, Malcovati et al report on the prevalence and clinical significance of mutations of SF3B1 in refractory anemia with ring sideroblasts (RARS) and refractory cytopenia with multilineage dysplasia and ring sideroblasts (RCMD-RS), defining a unique subset of myelodysplasis.1
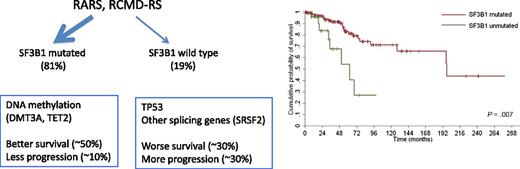
The majority of RARS and RCMD-RS have mutations in SF3B1, and these cases have a better outcome compared with cases without the mutation. SF3B1 mutants also have a high percentage of mutations involving DNA methylation genes. In wild-type SF3B1, those with mutations in other splicing genes also segregate with DNA methylation mutations, whereas those with no RNA splicing mutations have a higher prevalence of TP53 mutations. The figure has been adapted from data and Figure 2C in the article by Malcovati et al that begins on page 233.
The majority of RARS and RCMD-RS have mutations in SF3B1, and these cases have a better outcome compared with cases without the mutation. SF3B1 mutants also have a high percentage of mutations involving DNA methylation genes. In wild-type SF3B1, those with mutations in other splicing genes also segregate with DNA methylation mutations, whereas those with no RNA splicing mutations have a higher prevalence of TP53 mutations. The figure has been adapted from data and Figure 2C in the article by Malcovati et al that begins on page 233.
The myelodysplastic syndromes (MDSs) are a heterogeneous group of clonal malignancies characterized by cytopenias arising either de novo or after the genotoxic insult of prior radio- or chemotherapy. The morbidity and mortality of MDS stem from the complications of the low blood counts (infection and bleeding), or from progression to acute myeloid leukemia (AML). Alas, there are seemingly more classification schemes than effective therapies.2-4 Some classification systems focus on diagnosis using morphologic changes such as dysplasia and blast counts (World Health Organization [WHO]), whereas others focus on prognosis using clinical features such as blasts, cytogenetic karyotype, and extent of cytopenias (International Prognostic Scoring System [IPSS]). In sum, these classification systems allow for a prediction of the outcome. These predictions are not based on therapy, but because there are no great therapies in MDS, this distinction is unfortunately generally a moot point.
Similar classification systems in AML have recently incorporated certain mutation of certain genes (eg, FLT3 and NPM1) into schemes historically driven by cytogenetic findings. It would seem likely that similar refinement is on the horizon for MDS, as in the last few years the mutational landscape has been pursued and defined, with the characterization of functional categories of mutations, as well as the identification of certain genes associated with favorable, and poor, outcomes. Indeed, sequencing of genes established in myeloid malignancies first identified several genes associated with poor outcome in MDS (ASXL1, ETV6, EZH2, RUNX1, and TP53).5 With next-generation sequencing, these genes and novel genes were discovered. Similar to AML, a small set of mutations accounted for most recurrent mutations, and each patient had an average of <10 mutations, far less than in solid tumors. When clustered into biological pathways, genes involved in cell signaling (NRAS, JAK2), DNA methylation (TET2, DMT3A, IDH1/2), cohesion complex (RAD21), and chromatin regulation (ASXL1, EZH2) were characterized. In addition, 2 groups discovered a new pathway commonly mutated in MDS involved in RNA splicing.6-8
RNA splicing is a highly specific activity that allows eukaryotes to create an increased diversity of proteins coded from a limited set of genes via alternative splicing of messenger RNA (mRNA). Mutations in genes involved in the E/A splicing complex, essential in recognizing the 3′ splice site of mRNA, were found to occur in >50% of MDS cases, making the splicing the most common pathway mutated in MDS. The splicing pathway appears relatively intact in AML, but is also affected in a subset of chronic myelomonocytic leukemia cases. One of the most commonly mutated is splicing factor 3b, subunit 1 (SF3B1), occurring in roughly 25% of all MDS cases.
Curiously, mutations in SF3B1 appear to be predominately in MDS cases involving ringed sideroblasts, especially RARS and RCMD-RS. In this issue, Malcovati et al describe some of the clinical and genetic associations of SF3B1 in MDS cases with ringed sideroblasts.1 Previous to this publication, this group has done substantial work detailing the role of SF3B1 mutation in MDS. In short, they showed (1) a high prevalence of SF3B1 mutations in MDS cases characterized by ringed sideroblasts (65%),6 (2) a strong positive predictive value of SF3B1 of 98% for the ringed sideroblast phenotype (regardless of WHO category),9 and (3) a relatively favorable outcome of patients with the SF3B1 mutation.10
In the current manuscript, the authors expand our understanding of the role of SF3B1 in RARS and RCMD-RS. The authors studied 293 cases with >1% ringed sideroblasts (243 with MDS), with 159 cases having the RARS or RCMD-RS diagnoses. Of these cases, 81% had mutated SF3B1, and the presence of the mutation was associated with a substantially improved survival and lower rate of progression to AML compared with patients without the mutation. Moreover, SF3B1 mutations segregated with mutations of DNA methylation genes (TET2, DMT3A), whereas cases with ringed sideroblasts without SF3B1 were often associated with TP53 mutations (which may explain the outcome differences between the 2 groups; see figure). Of note, nearly 20% of SF3B1 wild-type cases had mutations in other RNA splicing genes, and these cases also often partnered with DNA methylation gene mutations.
Gene sequencing efforts in myeloid malignancies have largely charted the mutational “landscape.” This map allows us to (1) have some idea of the fundamental biology underlying the disease, (2) define potential drug targets, and (3) refine outcome expectations, especially when there are no “knockout” therapies (like in chronic myeloid leukemia). The consequence is also the further subclassification of myeloid malignancies, thus making relatively rare diseases into extremely rare ones. One obvious challenge is to cleverly design clinical studies given the myriad subcategories of disease. The higher bar is understanding the biology of how the various mutations and pathways merge to cause disease. The work Malcovati et al, along with the other fine studies noted above, gets us 1 step further down the road to cures.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal