Key Points
The platelet defect associated with Paris-Trousseau thrombocytopenia and Jacobsen syndrome is caused by an abnormal transcription factor FLI1.
FLI1 DNA–binding ETS domain mutations cause bleeding disorders with both autosomal dominant and recessive patterns of inheritance.
Abstract
Hemizygous deletion of a variable region on chromosome 11q containing FLI1 causes an inherited platelet-related bleeding disorder in Paris-Trousseau thrombocytopenia and Jacobsen syndrome. These multisystem disorders are also characterized by heart anomalies, changes in facial structure, and intellectual disability. We have identified a consanguineous family with autosomal recessive inheritance of a bleeding disorder that mimics Paris-Trousseau thrombocytopenia but has no other features of the 11q23 deletion syndrome. Affected individuals in this family have moderate thrombocytopenia; absent collagen-induced platelet aggregation; and large, fused α-granules in 1% to 5% of circulating platelets. This phenotype was caused by a FLI1 homozygous c.970C>T-point mutation that predicts an arginine-to-tryptophan substitution in the conserved ETS DNA–binding domain of FLI1. This mutation caused a transcription defect at the promoter of known FLI1 target genes GP6, GP9, and ITGA2B, as measured by luciferase assay in HEK293 cells, and decreased the expression of these target proteins in affected members of the family as measured by Western blotting of platelet lysates. This kindred suggests abnormalities in FLI1 as causative of Paris-Trousseau thrombocytopenia and confirms the important role of FLI1 in normal platelet development.
Introduction
Dominant inheritance of deletion 11q23 has been associated with a bleeding defect with large α-granules and abnormal megakaryocyte morphology in Paris-Trousseau thrombocytopenia (OMIM 188025). Patients with this disorder, and the closely associated Jacobsen syndrome, have variably sized chromosomal deletions associated with different components of a syndrome encompassing growth restriction and intellectual disability, bleeding with thrombocytopenia, and abnormal development of the heart and face. The platelet defect has been attributed to hemizygous deletion of the transcription factor FLI1; however, murine studies demonstrate no blood phenotype in mice with heterozygous Fli1 deletion, and other genes in the deleted region have been suggested to contribute to the pathogenesis.1-3
Methods
The kindred was identified from individuals participating in a human research ethics committee–approved study using a gene panel to identify mutations associated with macrothrombocytopenia. All individuals provided written informed consent and research was conducted in accordance with the Declaration of Helsinki.
Individuals were phenotyped with automated full blood count analysis, light transmission aggregometry or whole-blood impedance aggregometry (Multiplate), flow cytometry, and Western blotting. Bleeding scores were calculated using the BAT tool.4 Blood samples for electron microscopy were collected into ethylenediaminetetraacetic acid tubes. Genotyping was performed using Sanger sequencing and the Illumina Miseq platform as per the manufacturer’s instructions. In vitro luciferase studies were performed on HEK293 cells (see supplemental Methods, found on the Blood Web site).
Results and discussion
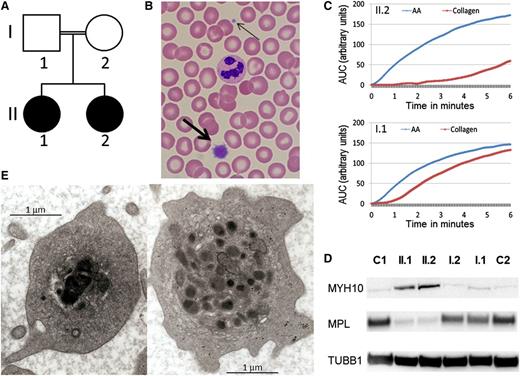
We report a consanguineous Caucasian kindred with the autosomal recessive inheritance of a bleeding disorder (Figure 1A). The parents are second cousins and have a normal automated full blood count (platelet count I.1: 227 × 109/L, I.2: 220 × 109/L) and blood film examination. They have no bleeding history (bleeding score 0 for both individuals) and no evidence of a functional platelet defect. In contrast, both adult children have a lifelong bleeding history of moderately severe mucosal bleeding and menorrhagia with abnormal bleeding scores of 12 for II.1 and 17 for II.2. Both affected individuals are moderately thrombocytopenic (II.1: 65 × 109/L and II.2: 76 × 109/L) with prominent large platelets on blood film examination (Figure 1B). Light transmission aggregometry revealed absence of collagen-induced platelet aggregation, reduced aggregation to adenosine diphosphate and epinephrine (primary wave only) but normal aggregation to arachidonic acid and ristocetin, a pattern seen in some dominant FLI1 mutations5 and Gray platelet syndrome.6 Multiplate testing demonstrated a marked decrease in collagen-induced platelet aggregation (Figure 1C), with normal response to arachidonic acid. By flow cytometry, without platelet size correction, platelet surface collagen receptor expression appeared normal, whereas platelet glycoprotein IIbIIIa complexes were reduced. After adenosine diphosphate activation, p-selectin and CD63 were normally expressed, but binding of the fibrinogen mimetic Pac-1 was reduced; platelet mepacrine uptake and release were normal (data not shown). MYH10, a protein associated with FLI1 and RUNX1 abnormalities,7 was detected in the platelets of the 2 probands by Western blotting, but the thrombopoietin receptor (MPL) was nearly absent (Figure 1D). Platelet electron microscopy of the 2 affected individuals showed some platelets with large, fused, and electron-dense α-granules characteristic of the Paris-Trousseau defect1,8 (Figure 1E). These large granules were present in ∼4% of circulating platelets (11/292), consistent with the previously reported frequency in families with Paris-Trousseau thrombocytopenia.1 Red cell and white cell automated blood parameters for both affected individuals were normal, and there were no other clinical features associated with Paris-Trousseau or Jacobsen syndromes.
Autosomal recessive inheritance of a platelet phenotype that mimics Paris-Trousseau thrombocytopenia. (A) Pedigree of the consanguineous family with the probands in black. (B) MGG stained blood film showing a normal sized platelet (thin arrow) and giant platelet (thick arrow). (C) Platelet aggregation measured by Multiplate in homozygous R324W FLI1 blood (top, II.2) and heterozygous R324W FLI1 (bottom, I.1) in response to arachidonic acid (AA) in blue and collagen in red. (D) Western blotting using monoclonal antibodies against MYH10 (mouse monoclonal, clone A-3) and MPL (rabbit polyclonal), with tubulin (mouse monoclonal, clone SAP.4G5) used as protein loading control, on probands (II.1 and II.2), their parents (I.1 and I.2), and unrelated controls (C1, C2). (E) Photographs of II.1 platelet EM demonstrating α-granule defect characteristic of Paris-Trousseau thrombocytopenia on the left compared with a platelet with normal granule appearance on the right.
Autosomal recessive inheritance of a platelet phenotype that mimics Paris-Trousseau thrombocytopenia. (A) Pedigree of the consanguineous family with the probands in black. (B) MGG stained blood film showing a normal sized platelet (thin arrow) and giant platelet (thick arrow). (C) Platelet aggregation measured by Multiplate in homozygous R324W FLI1 blood (top, II.2) and heterozygous R324W FLI1 (bottom, I.1) in response to arachidonic acid (AA) in blue and collagen in red. (D) Western blotting using monoclonal antibodies against MYH10 (mouse monoclonal, clone A-3) and MPL (rabbit polyclonal), with tubulin (mouse monoclonal, clone SAP.4G5) used as protein loading control, on probands (II.1 and II.2), their parents (I.1 and I.2), and unrelated controls (C1, C2). (E) Photographs of II.1 platelet EM demonstrating α-granule defect characteristic of Paris-Trousseau thrombocytopenia on the left compared with a platelet with normal granule appearance on the right.
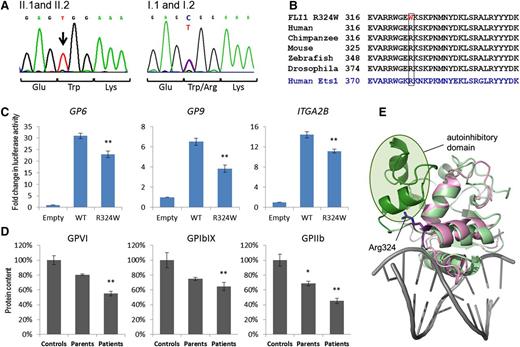
Coding exons of FLI1 were sequenced using Sanger and massively parallel sequencing. A homozygous c.970C>T mutation in exon 9 was identified in the 2 affected daughters and this mutation was present in the heterozygous state in both unaffected parents (Figure 2A). This base change predicts mutation of the conserved arginine in position 324 to a hydrophobic tryptophan in the DNA-binding loop between the α-2 and α-3 helices of the FLI1 ETS domain9 (Figure 2B). This mutation is predicted to be damaging by SIFT,10 Polyphen-2,11 and Mutation-Taster,12 and mutation of the adjacent Lys325 has previously been reported to result in loss of DNA binding.13,14
The FLI1 c.960C>T mutation in the ETS domain alters transcriptional activity of the protein. (A) Sequence traces showing the homozygous 970C>T mutation in patients II.1 and II.2 (left) and parents (right). (B) Amino acid homology in the ETS domain of FLI1 and ETS1. Arg324 is highly conserved among species. (C) HEK263 cells were transfected with an empty vector control, wild-type FLI1, or the mutant FLI1 c.970C>T transcript, and transcriptional activity was measured by the ratio of signal from the GP9, GP6, or ITGA2B promoter (firefly) compared with a control promoter (renilla). The data are expressed in fold change in luciferase activity and represent the mean ± standard error of the mean (N = 9) (*P < .05, **P < .01). (D) Western blotting of platelet lysates from affected (II.1, II.2) unaffected (I.1, I.2) family members, and unrelated controls representing 100%. Quantitation was performed using ImageLab software (*P < .05, **P < .01). (E) Ribbon representation of the structure of the FLI1 ETS domain in pink demonstrating the position of Arg324. Superimposed in green is the structure of the ETS1 ETS domain bound to DNA (gray). The N-terminal autoinhibitory region is shown in dark green and the core ETS domain is shown in light green.
The FLI1 c.960C>T mutation in the ETS domain alters transcriptional activity of the protein. (A) Sequence traces showing the homozygous 970C>T mutation in patients II.1 and II.2 (left) and parents (right). (B) Amino acid homology in the ETS domain of FLI1 and ETS1. Arg324 is highly conserved among species. (C) HEK263 cells were transfected with an empty vector control, wild-type FLI1, or the mutant FLI1 c.970C>T transcript, and transcriptional activity was measured by the ratio of signal from the GP9, GP6, or ITGA2B promoter (firefly) compared with a control promoter (renilla). The data are expressed in fold change in luciferase activity and represent the mean ± standard error of the mean (N = 9) (*P < .05, **P < .01). (D) Western blotting of platelet lysates from affected (II.1, II.2) unaffected (I.1, I.2) family members, and unrelated controls representing 100%. Quantitation was performed using ImageLab software (*P < .05, **P < .01). (E) Ribbon representation of the structure of the FLI1 ETS domain in pink demonstrating the position of Arg324. Superimposed in green is the structure of the ETS1 ETS domain bound to DNA (gray). The N-terminal autoinhibitory region is shown in dark green and the core ETS domain is shown in light green.
To validate the identified FLI1 mutation and confirm the relationship between FLI1 and the Paris-Trousseau platelet phenotype, further platelet Western blotting and in vitro luciferase assays were performed. In HEK293 cells, there was a significant decrease in transcriptional activity associated with the FLI1c.970C>T mutant transcript at the promoter of validated FLI1-target genes, GP615 (mutant 22.96 vs wild-type 30.96, P < .0001, N = 9), GP916 (3.82 vs 6.49, P < .001, N = 12), and ITGA2B16 (11.12 vs 14.37, P = .013, N = 9) (Figure 2C). This transcriptional change was consistent with the reduced expression of glycoprotein VI (34% reduction, P < .001), GPIbIX (32% reduction, P = .038), and GPIIbIIIa (42.8% reduction, P < .01) observed in platelet lysates derived from affected individuals II.1 and II.2 as measured by Western blotting (Figure 2D).
The FLI1 inheritance pattern in this family is intriguing and does not reflect the dominant inheritance pattern of the platelet defect in Paris-Trousseau thrombocytopenia. Both parents exhibit the DNA-binding mutation in the heterozygous state but have no observable platelet defect, no abnormality of α-granule morphology, and no increase in MYH10. Both children have characteristic platelet changes associated with Paris-Trousseau syndrome but exhibit autosomal recessive inheritance of this defect. This pattern of inheritance is consistent with the genetic mouse model of Fli1 deletion, where mice with a heterozygous Fli1 deletion (Fli1+/−) demonstrate no hematologic phenotype and have a normal tail bleeding time.17 Homozygous deletion of Fli1 in mice is embryonic-lethal, but Fli1−/− megakaryocytes derived from murine fetal liver have a proliferation and differentiation defect that is not present in megakaryocytes derived from Fli1+/− tissue.17 The abnormality we describe, p.Arg324Trp, is not embryonic-lethal as is unlikely to represent a null allele. Our luciferase experiments demonstrate some transcriptional activity for the mutant FLI1 construct, and protein modeling suggests Arg324 does not directly contact DNA. This arginine is conserved in the related ETS-domain protein ETS1 (Figure 2E), where this residue makes direct contacts with an N-terminal autoinhibitory domain (Figure 2E, dark green) that is known to regulate DNA-binding affinity through a transition between a folded and unfolded state and perhaps regulate interactions with other partner proteins.18 Although such an autoinhibitory domain has not yet been identified in FLI1, it is possible that a related mechanism is at play in the current situation. Recently, 3 FLI1 mutations have been reported5 that are predicted to disrupt the third α-helix of the ETS domain that directly binds to the DNA groove. These mutations appear to cause thrombocytopenia with bleeding in the heterozygous state, suggesting that these mutations may be more damaging to this FLI1–DNA interaction.
Single-cell studies in individuals with Paris-Trousseau thrombocytopenia suggest FLI1 may be monoallelically expressed in a proportion of megakaryocyte progenitors,3 with an absence of normal FLI1 transcript in this cell subpopulation being responsible for the abnormal megakaryocyte and platelet differentiation observed. The homozygous FLI1 c.970C>T mutation identified in our 2 individuals is likely to produce a hypofunctional allele that is present in all hematopoietic cells and that appears to cause the same platelet granule pathology. These data strongly suggest that abnormalities of FLI1 function are responsible for the complex platelet defect observed in Paris-Trousseau thrombocytopenia and Jacobsen syndrome, and further suggest that FLI1 is an important transcriptional regulator of platelet granule development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr John Hewson, Institute for Clinical Pathology and Medical Research Sydney, for his assistance with electron microscopy.
Authorship
Contribution: M.-C.M.-K. and S.G. carried out mutational screening and analyzed data; M.-C.M.-K. and L.B. performed Western blotting and luciferase assays and analyzed data; M.-C.M.-K. and Q.C. generated vectors; C.M.W. and D.J.R. enrolled patients, provided biological samples, and analyzed clinical data; J.P.M. provided analytical tools needed for 3D modeling; T.A.B., D.J.R., M.-C.M.-K., and L.B. processed the samples for E.M. and analyzed the images; and M.-C.M.-K. and W.S.S. designed research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marie-Christine Morel-Kopp, Northern Blood Research Centre, Kolling building Level 11, Royal North Shore Hospital, Reserve Rd, St Leonards NSW 2065, Australia; e-mail: marie-christine.kopp@sydney.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal