Key Points
Mutations in the Gardos channel, encoded by the KCNN4 gene, were identified in individuals from 2 hereditary xerocytosis kindreds.
These findings support recent data indicating the Gardos channel plays a role in normal erythrocyte volume homeostasis.
Abstract
Hereditary xerocytosis (HX; MIM 194380) is an autosomal-dominant hemolytic anemia characterized by primary erythrocyte dehydration. In many patients, heterozygous mutations associated with delayed channel inactivation have been identified in PIEZO1. This report describes patients from 2 well-phenotyped HX kindreds, including from one of the first HX kindreds described, who lack predicted heterozygous PIEZO1-linked variants. Whole-exome sequencing identified novel, heterozygous mutations affecting the Gardos channel, encoded by the KCNN4 gene, in both kindreds. Segregation analyses confirmed transmission of the Gardos channel mutations with disease phenotype in affected individuals. The KCNN4 variants were different mutations in the same residue, which is highly conserved across species and within members of the small-intermediate family of calcium-activated potassium channel proteins. Both mutations were predicted to be deleterious by mutation effect algorithms. In sickle erythrocytes, the Gardos channel is activated under deoxy conditions, leading to cellular dehydration due to salt and water loss. The identification of KCNN4 mutations in HX patients supports recent studies that indicate it plays a critical role in normal erythrocyte deformation in the microcirculation and participates in maintenance of erythrocyte volume homeostasis.
Introduction
The hereditary xerocytosis (HX) syndromes are a group of disorders associated with erythrocyte dehydration clinically manifest as mild to moderate hemolytic anemia. Dominantly inherited missense mutations in PIEZO1 have been identified in most HX patients, primarily located in the highly conserved COOH terminus.1-3 This report describes genetic studies in 2 HX kindreds who lack heterozygous mutations in PIEZO1, identifying mutations in the KCNN4 gene encoding the Gardos channel.
Study design
Patients
Targeted exon capture and whole-exome sequencing
Targeted regions of genomic DNA were captured using a NimbleGen SeqCap EZExome V2.0 solution-based capture system according to the manufacturer’s protocol. The captured, purified, and amplified libraries targeting the exomes from the HX patients were sequenced on a HiSeq Analyzer with paired-end sequencing at 75 bp read length. Sequencing reads were processed and analyzed as described previously.8
Exome sequence analysis
Fastq sequence reads were aligned to the human genome (hg19build37) using BWA mem 0.7.9a software.9 Variant analysis was performed using GATK analysis software v.3.1-1.10,11 Sequencing reads in the region of known insertion/deletions (indels) in the 1000 Genomes (August 4, 2010) database were realigned to reduce false positive variants.
Individual base quality scores were empirically recalibrated using covariate information in combination with the variation in dbSNP build 137 single-nucleotide polymorphisms (SNPs) and from 629 human genome sequences from the 1000 Genomes Project.12,13 Base alignment quality scores were determined then recalibrated with GATK to further reduce false-positive calls.14 SNPs and indels were called using the GATK HaplotypeCaller Bayesian algorithm for variant discovery and genotyping, which uses base qualities and allele counts to determine probabilities for called variants. Indels and novel SNPs were annotated using Annovar, which determines if a SNP or indel changes a protein sequence or splice site and provides a variety of predictions (including PolyPhen2, SIFT, Mutation Taster, and GERP) to assess the likely effect of a mutation on protein function.
Sanger sequencing
Sanger sequencing was performed on an Applied Biosystems 3130XL capillary sequencer.
Results and discussion
Exome sequencing and novel variant identification
Whole-exome sequencing data were aligned to the human genome (hg19) and analyzed. High mean coverage of >96% was achieved, and >93.7% of all targeted bases were read more than 10 times for the HX samples, respectively, which is more than sufficient to determine genotypes across the genome with high specificity. Genotypes for single-nucleotide and indel variants were called for HX and normal samples using the GATK Haplotype Caller and submitted to the Annovar annotation pipeline.
Candidate gene identification
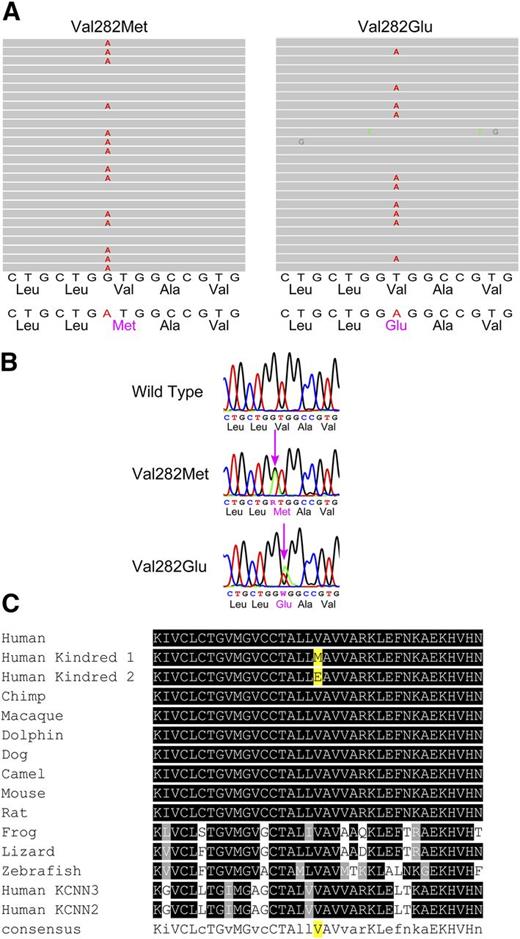
Assuming a model with dominant inheritance, we searched exome data for potential hydration-associated variants present in HX patients, but not in unaffected controls. In HX patients from kindred 1, a candidate was identified in the KCNN4 gene, with abnormal DNA sequences in the alignment of multiple short sequencing reads in exon 5 (Figure 1). The corresponding DNA sequence revealed a heterozygous G to A substitution (nucleotide position chr19:44273957) changing a Val (GTG) to Met (ATG) at amino acid 282 (NM_002250:c.G844A:p.V282M). We repeated the same analyses in the patient from kindred 2. A candidate variant was also identified in the KCNN4 gene, with abnormal DNA sequences in the alignment of multiple short sequencing reads in exon 5 (Figure 1). The corresponding DNA sequence revealed a heterozygous T to A substitution (nucleotide position chr19:44273956) changing a Val (GTG) to Glu (GAG) at amino acid 282 (NM_002250:c.T845A:p.V282E). Thus novel, nonsynonymous variants in the KCNN4 gene, which encodes the Gardos channel, were present in both kindreds, remarkably different mutations in the same amino acid.
Mutation identification. (A) Exome sequencing. Graphical representation of sequence tags from exome sequencing (exon 5) of a kindred 1 HX patient (left), where 17 of 31 full-length reads have an A instead of a G, leading to a missense mutation (Val to Met at amino acid 282), and a kindred 2 HX patient (right), where 41 of 84 full-length reads have an A instead of a T, leading to a missense mutation (Val to Glu at amino acid 282). (B) Sanger sequencing confirmation of KCNN4 gene mutations showing a partial exon 5 wild-type sequence (top), the corresponding sequence from a kindred 1 HX heterozygote (middle), and the corresponding sequence from a kindred 2 HX heterozygote (bottom). (C) Conservation of mutations across vertebrate species. The mutant amino acid residue identified in both HX patients is conserved across vertebrate species, including the clades of placental mammals, the extant Eutherians, and within members of the small-intermediate family of proteins.
Mutation identification. (A) Exome sequencing. Graphical representation of sequence tags from exome sequencing (exon 5) of a kindred 1 HX patient (left), where 17 of 31 full-length reads have an A instead of a G, leading to a missense mutation (Val to Met at amino acid 282), and a kindred 2 HX patient (right), where 41 of 84 full-length reads have an A instead of a T, leading to a missense mutation (Val to Glu at amino acid 282). (B) Sanger sequencing confirmation of KCNN4 gene mutations showing a partial exon 5 wild-type sequence (top), the corresponding sequence from a kindred 1 HX heterozygote (middle), and the corresponding sequence from a kindred 2 HX heterozygote (bottom). (C) Conservation of mutations across vertebrate species. The mutant amino acid residue identified in both HX patients is conserved across vertebrate species, including the clades of placental mammals, the extant Eutherians, and within members of the small-intermediate family of proteins.
No other deleterious variants were detected in the coding regions of the KCNN4 gene. The mutations in exon 5 of the KCNN4 gene identified by exome sequencing were validated by conventional Sanger sequencing (Figure 1). No heterozygous mutations were identified in the PIEZO1 gene locus.
Inheritance and mutation analyses
Segregation analyses confirmed transmission of the KCNN4 mutation with disease phenotype in affected individuals in kindred 1 (Figure 2). Mutations were absent in non-HX family members. The missense mutations were both predicted to be pathogenic by PolyPhen2, a tool that predicts possible impact of an amino acid substitution on the structure and function of proteins using physical and comparative considerations, SIFT, and several other mutation prediction algorithms. The mutations were both in conserved residues, with highly significant phastCons scores of 1.0 for both V282M and V282E and highly significant PhyloP conservation scores (V282M 7.489 and V282E 8.923). These amino acids are highly conserved across vertebrate species, including the clades of placental mammals, the extant Eutherians (Figure 1), and within members of the small-intermediate family of calcium-activated potassium channels. These variants are novel and are not present in the1000 Genomes, Exome Variant Server, or ExAC (>60 000 exomes) databases.
Cosegregation of KCNN4 gene mutations with HX phenotype. Patients in kindred 1 were assigned the diagnosis of HX (filled half circles) by clinical, laboratory, and biochemical methods.4-6 The KCNN4 gene mutation detected by exome sequencing cosegregated with disease phenotype in all affected HX individuals examined. Inheritance of the KCNN4 mutation is heterozygous.
Cosegregation of KCNN4 gene mutations with HX phenotype. Patients in kindred 1 were assigned the diagnosis of HX (filled half circles) by clinical, laboratory, and biochemical methods.4-6 The KCNN4 gene mutation detected by exome sequencing cosegregated with disease phenotype in all affected HX individuals examined. Inheritance of the KCNN4 mutation is heterozygous.
The amino acids affected by the KCNN4 missense mutations identified in the HX kindreds are located in predicted transmembrane loop S6 of the Gardos channel.
The Gardos channel is a heterotetrameric, voltage-independent potassium channel present in several cell types, including erythrocytes. It is activated by calcium, prostaglandin E, endothelin-1, and chemokines and inhibited by clotrimazole and other imidazole derivatives.15 In sickle erythrocytes, the Gardos channel is activated under deoxy conditions, leading to cellular dehydration due to salt and water loss.15,16 Senicapoc, a Gardos channel inhibitor, improved erythrocyte dehydration, numbers of dense cells, and hemolytic anemia in patients with sickle cell disease17 but did not decrease the rate of painful crises, and the study was ended early. It is tempting to consider Senicapoc for treatment of Gardos-linked HX.
Recent studies in normal erythrocytes demonstrated that local membrane deformation triggers a transient increase in calcium permeability with secondary activation of the Gardos channel and anionic channels, inducing significant dehydration during a brief deformation event in the microcirculation.18 These data suggest that erythrocytes change their shape and alter their volume during travel through the microvasculature.
While this report was in preparation, a report describing 2 HX kindreds with an identical mutation, Arg352His, was posted online.19 In both kindreds, KCNN4 genotype segregated with the HX phenotype, as observed in our kindred 1, fulfilling part of the definition of a new disease locus. A second critical part of the definition of a new disease gene locus is the identification of different mutations in the same gene locus. The combination of our 2 novel mutations with the online report’s single, different mutation provides compelling evidence that the KCNN4 gene is linked to the HX phenotype and fulfills criteria for a new disease gene locus.
Functional studies of HX-associated PIEZO1 mutations exhibit a partial gain-of-function phenotype with many mutants demonstrating delayed channel inactivation,2,20-22 suggesting increased cation permeability leads to erythrocyte dehydration. In KCNN4 mutant erythrocytes, preliminary data suggest altered channel activation kinetics also lead to erythrocyte dehydration.19
In normal patients, variation in indices of erythrocyte hydration is strongly influenced by genetic factors.23,24 Evidence continues to accumulate indicating that variants in genes encoding proteins involved in erythrocyte volume homeostasis contribute to their ability to ameliorate or worsen cellular hydration in patients with primary (HX) and secondary (sickle cell disease, thalassemia, and spherocytosis) disorders of erythrocyte hydration. The Gardos channel and its regulators are excellent candidates for these critical regulatory proteins.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr L. Michael Snyder, University of Massachusetts Medical Center, Worcester, MA, for patient referral and helpful discussions.
This work was supported in part by grants from the Doris Duke Foundation and the National Institutes of Health, National Institute of Diabetes and Digestive Diseases (DK104046).
Authorship
Contribution: E.G. designed and performed experiments and analyzed data; K.L.-G. designed and performed experiments; Y.M. designed and performed experiments; V.P.S. analyzed data; and P.G.G. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick G. Gallagher, Departments of Pediatrics, Pathology, and Genetics, Yale University School of Medicine, 333 Cedar St, PO Box 208064, New Haven, CT 06520-8064; e-mail: patrick.gallagher@yale.edu.