Key Points
miR-142 is an essential regulator of lymphocyte ontogenesis and is required for the generation of humoral and cellular immunity in mice.
miR-142-3p regulates B-cell homeostasis by controlling expression of BAFF-R.
Abstract
MicroRNAs (miRNAs) are a class of powerful posttranscriptional regulators implicated in the control of diverse biological processes, including regulation of hematopoiesis and the immune response. To define the biological functions of miR-142, which is preferentially and abundantly expressed in immune cells, we created a mouse line with a targeted deletion of this gene. Our analysis of miR-142−/− mice revealed a critical role for this miRNA in the development and homeostasis of lymphocytes. Marginal zone B cells expand in the knockout spleen, whereas the number of T and B1 B cells in the periphery is reduced. Abnormal development of hematopoietic lineages in miR-142−/− animals is accompanied by a profound immunodeficiency, manifested by hypoimmunoglobulinemia and failure to mount a productive immune response to soluble antigens and virus. miR-142−/− B cells express elevated levels of B-cell–activating factor (BAFF) receptor (BAFF-R) and as a result proliferate more robustly in response to BAFF stimulation. Lowering the BAFF-R gene dose in miR-142−/− mice rescues the B-cell expansion defect, suggesting that BAFF-R is a bona fide miR-142 target through which it controls B-cell homeostasis. Collectively, our results uncover miR-142 as an essential regulator of lymphopoiesis, and suggest that lesions in this miRNA gene may lead to primary immunodeficiency.
Introduction
Control of gene expression by microRNAs (miRNAs) has recently emerged as a key mechanism that regulates multiple aspects of immune homeostasis, from cell fate determination during hematopoiesis to fine-tuning of immune response to infection.1,2 Support for this notion was first provided by studies that investigated the consequences of global abrogation of miRNA processing in B and T lymphocytes.
Conditional deletion of Dicer, which encodes an endonuclease required for miRNA biogenesis, in CD19+ B cells drastically alters differentiation of peripheral B-cell subsets and results in expansion of marginal zone (MZ) B cells at the expense of follicular (FO) B cells.3 Moreover, germinal center (GC) formation, generation of long-lived plasma cells, and production of high-affinity class-switched antibodies in response to antigen challenge are impaired in mice with specific Dicer ablation in activated B cells.4
miRNA control is also indispensible for T-lymphocyte homeostasis and function, as T-cell–specific Dicer knockout (KO) mice display smaller peripheral T-cell compartments, as well as defective proliferation and cytokine production by helper T cells.5 Although the above-mentioned studies using global ablation of miRNA expression have clearly established an essential role for miRNAs in hematopoiesis and immunity, the challenge today is to determine the contribution of individual miRNA genes to the regulation of immunologic processes and to define the molecular mechanism by which they elicit this control.
miR-142 is one of a few miRNA genes that are preferentially expressed in cells of hematopoietic origin.6-8 It encodes 2 mature miRNA molecules (miR-142-3p and miR-142-5p) that are derived from opposite strands of a hairpin-like precursor. Both miRNAs are evolutionarily conserved and found in vertebrate genomes from fish to humans.
The biological role of miR-142 remains poorly understood. Knockdown of miR-142-3p expression in zebrafish revealed a critical role for this miRNA in hematopoiesis during embryonic development, hematopoietic stem cell homeostasis, neutrophil differentiation, and heart development.9-11 In mice, miR-142 is essential for normal development and function of megakaryocytes, dendritic, and mast cells.12-14
Our findings from miR-142–deficient mice suggest that this miRNA is necessary for lymphocyte homeostasis, with genetic ablation causing aberrant development of MZ B cells, B1 B cells, and peripheral T cells. Furthermore, we demonstrate that abrogation of miR-142 expression results in a combined immunodeficiency and extend our analysis to implicate miR-142-3p as the functional strand regulating the mature B-cell gene expression program. Finally, we provide genetic evidence that miR-142-3p may act through B-cell–activating factor receptor (BAFF-R) to control B-cell homeostasis. Collectively, our results establish miR-142 as a key regulator of mammalian lymphopoiesis and adaptive immunity, highlighting the importance of posttranscriptional regulation in these processes.
Methods
See supplemental Methods (available at the Blood Web site) for additional methods.
Generation of miR-142−/− mice
miR-142 null mice were generated by deleting ∼900 bp of genomic sequence that encompasses the miR-142 precursor (supplemental Figure 1A). A targeting vector containing 2 LoxP sites, a neomycin-positive selection cassette flanked by FRT sites, and a diphtheria toxin A–negative selection cassette was electroporated into C57BL/6J embryonic stem (ES) cells. The targeted ES cells were microinjected into host C57BL/6J-Tyrc-2J/J blastocysts and implanted into pseudopregnant foster mothers to produce chimeric mice. The resulting chimeras were bred with C57BL/6J mice to obtain offspring with the targeted miR-142 locus in the germline (F1 generation). F1 mice were crossed with a Cre-deleter strain (C57BL/6Tg(EIIa-Cre)) to remove the miR-142 locus and the neomycin cassette. The resulting miR-142+/− animals were interbred to produce the miR-142−/− line. All experimental procedures described in this study used C57BL/6J mice (obtained from The Jackson Laboratory and denoted hereafter as wild-type [WT]) for controls. All animal experiments were approved by the Institutional Animal Care and Use Committee of the City of Hope.
Bone marrow adoptive transfer
To perform competitive bone marrow (BM) reconstitution assays, 3 × 105 BM cells derived from WT C57BL/6 (CD45.2-positive) or miR-142−/− (CD45.2-positive) mice were cotransplanted with equal number of WT C57BL/6 (CD45.1-positive) BM cells into lethally irradiated (1000 rad) C57BL/6 (CD45.1) mice. Development of different hematopoietic lineages was analyzed by flow cytometry 12 to 17 weeks posttransplantation.
Global gene expression profiling using DNA microarrays
B cells from 6-week-old WT and miR-142 KO males (n = 3 per genome) were purified from single-cell suspensions of splenocytes using anti-mouse CD19 magnetic beads (Miltenyi Biotec) according to manufacturer’s protocol. Total RNA was isolated using a miRNeasy kit (QIAGEN) and subjected to analysis on a GeneChip Mouse Genome 430A 2.0 Array (Affymetrix). Differential analysis of gene expression in WT and miR-142 KO cells was performed using Partek’s Genomics Suite software. Analysis of enrichment of miR-142-3p and miR-142-5p seed sequences in expression profiles was performed by the Web-based SylArray software algorithm (http://www.ebi.ac.uk/enright-srv/sylarray/). DNA microarray and sample annotation data were deposited in the Gene Expression Omnibus under the accession number GSE61919.
Results
Immunoproliferative disorder in miR-142−/− mice
To test experimentally the physiological role of miR-142, we generated a mouse line with a targeted deletion of this miRNA gene (supplemental Figure 1A-B). As predicted, miR-142−/− mice display virtually no expression of both mature miR-142-3p and miR-142-5p in immune cells (supplemental Figure 1C).
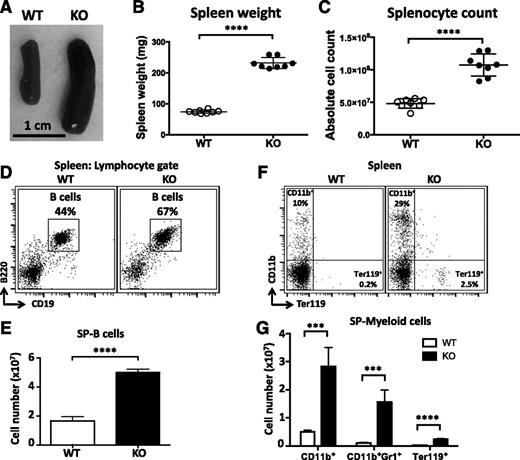
miR-142−/− mice are born at the expected Mendelian ratios and appear healthy and fertile. Gross morphologic analysis of miR-142−/− animals at necropsy did not reveal any apparent internal organ defects, except for significant enlargement of the spleen (Figure 1A-B). Splenomegaly was highly penetrant in miR-142−/− mice but was not accompanied by concurrent lymphadenopathy. Spleen enlargement was characterized by a marked increase in organ cellularity (Figure 1C), suggesting that miR-142 deletion results in an immunoproliferative disorder. Analysis of miR-142−/− splenocytes by flow cytometry revealed a significant expansion of both B-cell and myeloid cell populations (Figure 1D-G). The number of CD19+B220+ B lymphocytes in the KO spleen increased ∼2.5 times in comparison to the WT control (Figure 1E).
Immunoproliferative phenotypes in miR-142−/− mice. (A) Splenomegaly in miR-142−/− mice. Representative images of spleens from WT and KO mice. Spleen weights (B) and total splenocyte counts (C) in WT (n = 8) and miR-142−/−(n = 8) animals. (D-E) Enlargement of the B-cell compartment in the KO spleen. (D) Fluorescence-activated cell sorter (FACS) analysis of lymphocytes in WT and KO spleens with anti-B220 and anti-CD19 specific antibodies. B cells (B220+CD19+) are gated, and numbers indicate the percentage of cells in the gate. (E) Total B-cell counts in WT (n = 4) and KO (n = 4) spleens. (F-G) Myeloproliferation in the KO spleen. (F) FACS analysis of myeloid (CD11b+) and erythroid (Ter119+) lineages in WT and KO spleens with anti-CD11b and anti-Ter119 specific antibodies. Numbers indicate the percentage of cells in the quadrants. (G) Total counts of CD11b+, CD11b+GR1+, and Ter119+ cells in WT (n = 4) and KO (n = 4) spleens. Results are shown as means ± standard deviation (SD) and are representative of at least 3 experiments. P values were calculated using Student t test. ***P ≤ .001; ****P ≤ .0001. SP, spleen.
Immunoproliferative phenotypes in miR-142−/− mice. (A) Splenomegaly in miR-142−/− mice. Representative images of spleens from WT and KO mice. Spleen weights (B) and total splenocyte counts (C) in WT (n = 8) and miR-142−/−(n = 8) animals. (D-E) Enlargement of the B-cell compartment in the KO spleen. (D) Fluorescence-activated cell sorter (FACS) analysis of lymphocytes in WT and KO spleens with anti-B220 and anti-CD19 specific antibodies. B cells (B220+CD19+) are gated, and numbers indicate the percentage of cells in the gate. (E) Total B-cell counts in WT (n = 4) and KO (n = 4) spleens. (F-G) Myeloproliferation in the KO spleen. (F) FACS analysis of myeloid (CD11b+) and erythroid (Ter119+) lineages in WT and KO spleens with anti-CD11b and anti-Ter119 specific antibodies. Numbers indicate the percentage of cells in the quadrants. (G) Total counts of CD11b+, CD11b+GR1+, and Ter119+ cells in WT (n = 4) and KO (n = 4) spleens. Results are shown as means ± standard deviation (SD) and are representative of at least 3 experiments. P values were calculated using Student t test. ***P ≤ .001; ****P ≤ .0001. SP, spleen.
Abrogation of miR-142 expression results in defective differentiation and maturation of B cells
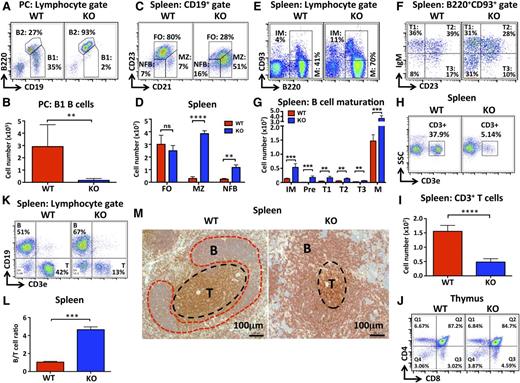
To probe further the role of miR-142 in B lymphocyte differentiation, we examined the ontogenesis of several B-cell subsets in miR-142−/− mice. Mammalian B cells are typically divided into 3 main subgroups based on functionality and localization: B1 B cells, MZ B cells, and FO B cells.15 We observed that the peritoneal cavities of the KO mice were virtually devoid of CD19+B220lo lymphocytes (Figure 2A-B), a cellular population that is typically defined as B1 B cells.16 Subsequent analysis of miR-142−/− peritoneal cavity lymphocytes using immunoglobulin M (IgM), CD11b, and CD5 markers further confirmed a marked contraction of the B1 B-cell population (supplemental Figure 2A-B). Deletion of miR-142 diminished production of both CD5+ B1a and CD5− B1b B cells, with a greater effect observed in the B1a subset (supplemental Figure 2A-B).
Deletion of miR-142 results in abnormal development of lymphoid cells. (A-B) Dramatic decrease in the number of B1 B cells in miR-142−/− mice. (A) FACS analysis of lymphocytes in the peritoneal cavity (PC) of WT and KO mice with anti-B220 and anti-CD19 specific antibodies. B1 (CD19+B220lo) and B2 (CD19+B220hi) cell populations are gated, and numbers indicate the percentage of cells in the gate. (B) Total B1 B-cell numbers in the PC of WT (n = 5) and KO (n = 6) mice. (C-D) Increased numbers of MZ-like B cells in miR-142−/− spleens. (C) FACS analysis of CD19+ B cells in WT and KO spleens with anti-CD21 and anti-CD23 antibodies. FO (CD21intCD23+), MZ (CD21hiCD23lo), and newly formed (NFB) (CD21intCD23lo) B-cell subsets are gated, and numbers indicate the percentage of cells in the gate. (D) Total numbers of FO, MZ, and NFB B cells in WT (n = 3) and KO (n = 3) spleens. (E-G) Altered B-cell maturation in miR-142−/− mice. (E) FACS analysis of WT and KO splenocytes with anti-B220 and anti-CD93 antibodies. Cells were pregated on lymphocytes. Immature (IM) (B220+CD93+) and mature (M) (B220+CD93−) B-cell populations are gated, and the numbers indicate the percentage of cells in the gate. (F) FACS analysis of B220+CD93+ immature B cells from WT and KO spleens with anti-IgM and anti-CD23 antibodies. Transitional T1 (IgM+CD23−), T2 (IgM+CD23+), T3 (IgM-CD23+), and precursor (pre-B) (IgM−CD23−) B cells are gated, and numbers indicate the percentage of cells in the gate. (G) Total numbers of pre-B, immature, transitional (T1-T3), and mature B cells in WT (n = 4) and KO (n = 4) spleens. (H-J) Decrease in peripheral T-cell numbers, but normal central T cell development in miR-142−/− mice. (H) FACS analysis of splenocytes in WT and KO mice with anti-CD3ε antibodies. T cells (CD3+) are gated, and numbers indicate the percentage of cells in the gate. (I) Total number of T cells in WT (n = 8) and KO (n = 8) spleens. (J) FACS analysis of WT and KO thymocytes with anti-CD4 and anti-CD8 antibodies. Numbers indicate the percentage of cells in the quadrants. (K-M) Elevated B-/T-cell ratio in miR-142−/− mice. (K) FACS analysis of WT and KO splenocytes with anti-CD3ε and anti-CD19 antibodies. B cells (CD19+) and T cells (CD3+) are gated, and numbers indicate the percentage of cells in the gate. (L) The ratio of B to T cells in WT (n = 3) and KO (n = 3) spleens. (M) Immunohistochemical analysis of spleen sections from WT and miR-142 KO mice with anti-B220 (red stain) and anti-CD3 (brown stain) antibodies. Scale bar, 100 μm. Note abnormal spleen architecture in the KO: smaller T-cell zone and less organized, less compact B-cell zone. Results are shown as means ± SD and are representative of at least 3 independent experiments. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. B, B-cell follicle; ns, not significant; T, T-cell zone.
Deletion of miR-142 results in abnormal development of lymphoid cells. (A-B) Dramatic decrease in the number of B1 B cells in miR-142−/− mice. (A) FACS analysis of lymphocytes in the peritoneal cavity (PC) of WT and KO mice with anti-B220 and anti-CD19 specific antibodies. B1 (CD19+B220lo) and B2 (CD19+B220hi) cell populations are gated, and numbers indicate the percentage of cells in the gate. (B) Total B1 B-cell numbers in the PC of WT (n = 5) and KO (n = 6) mice. (C-D) Increased numbers of MZ-like B cells in miR-142−/− spleens. (C) FACS analysis of CD19+ B cells in WT and KO spleens with anti-CD21 and anti-CD23 antibodies. FO (CD21intCD23+), MZ (CD21hiCD23lo), and newly formed (NFB) (CD21intCD23lo) B-cell subsets are gated, and numbers indicate the percentage of cells in the gate. (D) Total numbers of FO, MZ, and NFB B cells in WT (n = 3) and KO (n = 3) spleens. (E-G) Altered B-cell maturation in miR-142−/− mice. (E) FACS analysis of WT and KO splenocytes with anti-B220 and anti-CD93 antibodies. Cells were pregated on lymphocytes. Immature (IM) (B220+CD93+) and mature (M) (B220+CD93−) B-cell populations are gated, and the numbers indicate the percentage of cells in the gate. (F) FACS analysis of B220+CD93+ immature B cells from WT and KO spleens with anti-IgM and anti-CD23 antibodies. Transitional T1 (IgM+CD23−), T2 (IgM+CD23+), T3 (IgM-CD23+), and precursor (pre-B) (IgM−CD23−) B cells are gated, and numbers indicate the percentage of cells in the gate. (G) Total numbers of pre-B, immature, transitional (T1-T3), and mature B cells in WT (n = 4) and KO (n = 4) spleens. (H-J) Decrease in peripheral T-cell numbers, but normal central T cell development in miR-142−/− mice. (H) FACS analysis of splenocytes in WT and KO mice with anti-CD3ε antibodies. T cells (CD3+) are gated, and numbers indicate the percentage of cells in the gate. (I) Total number of T cells in WT (n = 8) and KO (n = 8) spleens. (J) FACS analysis of WT and KO thymocytes with anti-CD4 and anti-CD8 antibodies. Numbers indicate the percentage of cells in the quadrants. (K-M) Elevated B-/T-cell ratio in miR-142−/− mice. (K) FACS analysis of WT and KO splenocytes with anti-CD3ε and anti-CD19 antibodies. B cells (CD19+) and T cells (CD3+) are gated, and numbers indicate the percentage of cells in the gate. (L) The ratio of B to T cells in WT (n = 3) and KO (n = 3) spleens. (M) Immunohistochemical analysis of spleen sections from WT and miR-142 KO mice with anti-B220 (red stain) and anti-CD3 (brown stain) antibodies. Scale bar, 100 μm. Note abnormal spleen architecture in the KO: smaller T-cell zone and less organized, less compact B-cell zone. Results are shown as means ± SD and are representative of at least 3 independent experiments. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. B, B-cell follicle; ns, not significant; T, T-cell zone.
In the spleen, miR-142−/− B2 B lymphocytes displayed dramatic changes in the expression of CD21 and CD23 surface markers. CD23 expression was downregulated, whereas the level of CD21 increased (Figure 2C). As a result, production of CD21hiCD23− MZ-like B cells was elevated in miR-142−/− mice (Figure 2C-D). In contrast, the frequency of the CD21midCD23+ FO B-cell population decreased; however, the total number of FO B cells in miR-142−/− spleens, although trending lower, was not statistically different (Figure 2C-D and supplemental Figure 5B). MZ B cells normally express high surface levels of the CD1d antigen, which is believed to participate in the presentation of lipid antigens to natural killer T cells.17 In agreement with such observations, we found a significant increase in the number of B220+CD21hiCD1dhi cells in the KO spleen (supplemental Figure 2C). Expansion of the MZ-like B-cell population in miR-142−/− mice was further confirmed by immunohistochemical analysis of spleen sections stained with anti-IgM and anti-mucosal vascular addressin cell adhesion molecule 1 (MAdCam1) antibodies (supplemental Figure 2D). We formally measured the areas of MZ and FO B-cell regions in WT and miR-142−/− lymphoid follicles. In KO mice, we observed that the average MZ size increased ∼2.4-fold, whereas the average FO zone area contracted (supplemental Figure 2E-F). Consequently, miR-142−/− spleens displayed an approximately fourfold increase in the ratio between MZ and FO areas (supplemental Figure 2G). The total number of CD19+ B cells in miR-142−/− lymph nodes (LNs) was reduced, although their relative frequency was not altered (supplemental Figure 2J,M). Because mouse LNs are devoid of the MZ structure, these results are consistent with a decrease in FO B cells. In summary, several independent lines of evidence support the notion that deletion of miR-142 results in the expansion of MZ-like B cells. Interestingly, miR-142-3p expression in FO B cells is relatively more abundant in comparison with MZ B cells (Figure 6B), suggesting that lower expression of this miRNA may favor development toward the MZ B-cell lineage.
Our analysis of B-cell maturation in miR-142−/− spleen revealed a significant increase in both mature (B220+CD93−) and immature (B220+CD93+) populations (Figure 2E,G). Further examination of immature B cells with anti-IgM and anti-CD23 antibodies found little change in the frequency of transitional (T1-T3) B cells; however, the absolute number of cells in all 3 transitional stages was significantly increased (Figure 2F,G). We also observed an abnormal accumulation of CD23−IgM− B-cell precursors in miR-142−/− spleens, suggesting that miR-142 deletion perturbs early B-cell development. However, extended analysis of the miR-142 role in early B-cell development is beyond the scope of this study and will be published elsewhere. A higher frequency of immature B cells was also observed in miR-142−/− LNs (supplemental Figure 2N,O). Taken together, our findings indicate that miR-142 negatively regulates B2 B-cell differentiation and output.
miR-142 deletion reduces T-cell abundance in the periphery
In contrast to B-cell accumulation, our analysis of miR-142−/− splenocytes revealed a strong reduction in the number of CD3+ T cells (Figure 2H-I). Deletion of miR-142 resulted in an approximately threefold drop in splenic T-cell numbers (Figure 2I), but had little effect on the ratio between CD4+ and CD8+ T cells (supplemental Figure 2H-I). The total number of CD3+ T cells was also reduced in miR-142−/− LNs (supplemental Figure 2J,L). At the same time, thymic development of T lymphocytes appeared normal in miR-142−/− mice (Figure 2J), suggesting that miR-142 controls either survival or homing of mature T cells to the peripheral lymphoid organs.
Overall, because of the expanding B-cell compartment and declining T-cell numbers, the B- to T-cell ratio in KO spleens was strongly skewed toward B cells (Figure 2K-L). For each T lymphocyte in the KO spleen, we observed ∼5 B cells, whereas the WT mice displayed an almost 1-to-1 ratio (Figure 2L). In agreement with these results, immunohistochemical analysis of the KO spleen sections stained with anti-B220 and anti-CD3 antibodies revealed a sharply diminished T-cell zone and marked enlargement of the B-cell area in lymphoid follicles (Figure 2M and supplemental Figure 2F). Moreover, the B-cell zone in KO spleens lost its normal compact organization and became highly disorganized in shape.
Aberrant development of lymphocytes in miR-142−/− mice is cell intrinsic
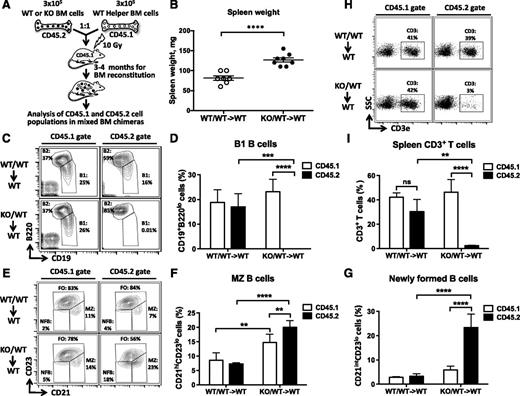
To determine whether defective ontogenesis of lymphocytes in the KO mice is the result of ablation of miR-142 expression in immune cells and not in stromal cells, we performed mixed BM transplantation experiments. Lethally irradiated C57BL/6J mice were injected with miR-142−/− BM cells together with an equal number of WT BM helper cells (Figure 3A). Gross morphologic examination of resulting chimeric mice 12 to 17 weeks post–BM transplantation revealed moderate splenomegaly in animals that received miR-142−/− BM (KO/WT→WT), but not in the control group (WT/WT→WT) (Figure 3B). Similar to our observations with miR-142−/− animals, KO/WT→WT spleens were more cellular than the control organs (supplemental Figure 3). Our analysis of chimeric lymphocyte populations by flow cytometry using 2 isoforms of the CD45 antigen to trace cells of WT (CD45.1/2) and KO (CD45.2) origin revealed several defects. First, we found no CD45.2+ B1 B cells in the peritoneal cavities of KO/WT→WT animal cohort, whereas CD45.1+ B1 B cells were thriving in the same mice (Figure 3C-D). Second, in KO/WT→WT spleens, the CD45.2+ newly formed, immature B-cell population (CD21intCD23lo) was ∼4 times bigger than a similar coexisting CD45.1+ population (Figure 3E,G). In addition, we found an expansion of MZ-like B cells (B220+CD21hiCD23−) in both CD45.1+ and CD45.2+ compartments in KO/WT→WT, but not WT/WT→WT chimeras (Figure 3E-F). This result implies that miR-142−/− immune cells can somehow (most likely by secreting a protein factor) drive differentiation of WT B cells toward the MZ lineage. Finally, KO/WT→WT spleens displayed a marked decrease in the number of CD45.2+ T cells, whereas the number of CD45.1+ CD3+ cells did not change significantly (Figure 3H-I). Taken together, our results strongly suggest that aberrant lymphocyte ontogenesis in miR-142−/− mice was cell autonomous in nature.
miR-142 functions in a cell-autonomous manner to control the development of B and T lymphocytes. (A) Diagram of mixed BM chimera generation. WT or miR-142−/− (CD45.2+) BM cells were mixed at 1:1 ratio with helper WT (CD45.1+) BM cells and transplanted into lethally irradiated WT (CD45.1+) hosts. Resulting chimeric mice were analyzed 12-17 weeks posttransplantation. (B) Splenomegaly in WT mice receiving miR-142−/− BM. Spleen weight in mice reconstituted with a mixed WT BM (WT/WT→WT) or a mixture of WT and KO BM cells (KO/WT→WT). (C-D) B1 B cell differentiation defect in mixed miR-142−/− BM chimeras. (C) FACS analysis of lymphocytes in peritoneal cavity of mixed BM chimeric mice with anti-B220 and anti-CD19 specific antibodies. B1 (CD19+B220lo) and B2 (CD19+B220hi) cell populations are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (D) Percentage of B1 B cells in peritoneal cavity of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (E-G) Excessive production of MZ and NFB B cells in mixed KO/WT→WT BM chimeras. (E) FACS analysis of B220+ B lymphocytes in the spleen of mixed BM chimeric mice with anti-CD21 and anti-CD23 specific antibodies. FO (CD21intCD23+), MZ (CD21hiCD23lo), and NFB (CD21intCD23lo) B-cell subsets are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (F) Percentage of MZ B cells in the spleens of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (G) Percentage of NFB cells in spleens of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (H-I) Decrease in the number of peripheral T cells in the mixed miR-142−/− BM chimeras. (H) FACS analysis of lymphocytes in the spleens of mixed BM chimeric mice with anti-CD3ε specific antibodies. T cells (CD3+) are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (I) Percentage of CD3+ T cells in the spleens of WT/WT→WT (n = 3) and KO/WT→WT (n = 3) mixed BM chimeric mice. Results are shown as means ± SD and are representative of 2 independent experiments. P values were calculated using either uncorrected Fisher’s least significant difference test or Student t test. ns, not significant; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. NF, newly formed.
miR-142 functions in a cell-autonomous manner to control the development of B and T lymphocytes. (A) Diagram of mixed BM chimera generation. WT or miR-142−/− (CD45.2+) BM cells were mixed at 1:1 ratio with helper WT (CD45.1+) BM cells and transplanted into lethally irradiated WT (CD45.1+) hosts. Resulting chimeric mice were analyzed 12-17 weeks posttransplantation. (B) Splenomegaly in WT mice receiving miR-142−/− BM. Spleen weight in mice reconstituted with a mixed WT BM (WT/WT→WT) or a mixture of WT and KO BM cells (KO/WT→WT). (C-D) B1 B cell differentiation defect in mixed miR-142−/− BM chimeras. (C) FACS analysis of lymphocytes in peritoneal cavity of mixed BM chimeric mice with anti-B220 and anti-CD19 specific antibodies. B1 (CD19+B220lo) and B2 (CD19+B220hi) cell populations are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (D) Percentage of B1 B cells in peritoneal cavity of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (E-G) Excessive production of MZ and NFB B cells in mixed KO/WT→WT BM chimeras. (E) FACS analysis of B220+ B lymphocytes in the spleen of mixed BM chimeric mice with anti-CD21 and anti-CD23 specific antibodies. FO (CD21intCD23+), MZ (CD21hiCD23lo), and NFB (CD21intCD23lo) B-cell subsets are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (F) Percentage of MZ B cells in the spleens of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (G) Percentage of NFB cells in spleens of WT/WT→WT (n = 4) and KO/WT→WT (n = 4) mixed BM chimeric mice. (H-I) Decrease in the number of peripheral T cells in the mixed miR-142−/− BM chimeras. (H) FACS analysis of lymphocytes in the spleens of mixed BM chimeric mice with anti-CD3ε specific antibodies. T cells (CD3+) are gated and numbers indicate the percentage of cells in the gate. Cells were first gated on the basis of CD45.1 (left panel) or CD45.2 (right panel) expression. (I) Percentage of CD3+ T cells in the spleens of WT/WT→WT (n = 3) and KO/WT→WT (n = 3) mixed BM chimeric mice. Results are shown as means ± SD and are representative of 2 independent experiments. P values were calculated using either uncorrected Fisher’s least significant difference test or Student t test. ns, not significant; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. NF, newly formed.
Hypoimmunoglobulinemia in miR-142−/− mice
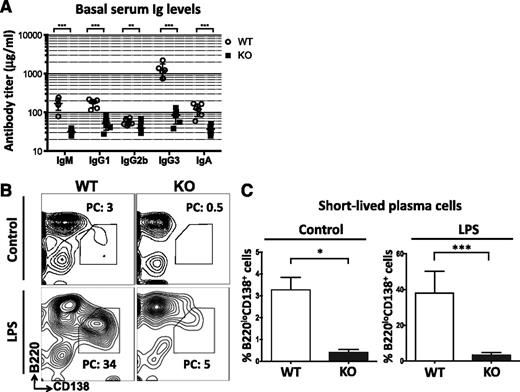
The presence of multiple B cell defects in miR-142−/− mice prompted us to examine the steady-state level of antibody production in these animals. Surprisingly, despite a significant expansion of the B-cell compartment, we found a strong decrease in the levels of all tested serum immunoglobulins in unimmunized miR-142−/− mice (Figure 4A). In addition, the ability of miR-142−/− B cells to differentiate in vitro into B220loCD138+ short-lived plasma cells18 in response to bacterial lipopolysaccharide was greatly diminished (Figure 4B-C). Our findings suggest that the B cells accumulating in miR-142−/− mice are functionally impaired.
Hypogammaglobulinemia and defective B-cell function in miR-142−/− mice. (A) Hypoimmunoglobulinemia in miR-142−/− mice. Analysis of serum immunoglobulins (Igs) in 8-week-old female WT (n = 6) and KO (n = 6) mice by enzyme-linked immunosorbent assay (ELISA). (B-C) miR-142 KO B cells have a significantly reduced capacity to differentiate into short-lived plasma cells. (B) FACS analysis of WT and KO splenocytes either left unstimulated or stimulated with lipopolysaccharide (20 μg/mL) for 3 days. Short-lived plasma cells (B220loCD138+) are gated and numbers indicate the percentage of cells in the gate. (C) Percentage of B220loCD138+ plasmablasts in unstimulated (n = 4) or lipopolysaccharide -stimulated (n = 6) WT and KO splenocyte cultures. Results are shown as means ± SD and are representative of 2 independent experiments. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001. LPS, lipopolysaccharide; PC, plasma cells.
Hypogammaglobulinemia and defective B-cell function in miR-142−/− mice. (A) Hypoimmunoglobulinemia in miR-142−/− mice. Analysis of serum immunoglobulins (Igs) in 8-week-old female WT (n = 6) and KO (n = 6) mice by enzyme-linked immunosorbent assay (ELISA). (B-C) miR-142 KO B cells have a significantly reduced capacity to differentiate into short-lived plasma cells. (B) FACS analysis of WT and KO splenocytes either left unstimulated or stimulated with lipopolysaccharide (20 μg/mL) for 3 days. Short-lived plasma cells (B220loCD138+) are gated and numbers indicate the percentage of cells in the gate. (C) Percentage of B220loCD138+ plasmablasts in unstimulated (n = 4) or lipopolysaccharide -stimulated (n = 6) WT and KO splenocyte cultures. Results are shown as means ± SD and are representative of 2 independent experiments. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001. LPS, lipopolysaccharide; PC, plasma cells.
Combined immunodeficiency in miR-142−/− mice
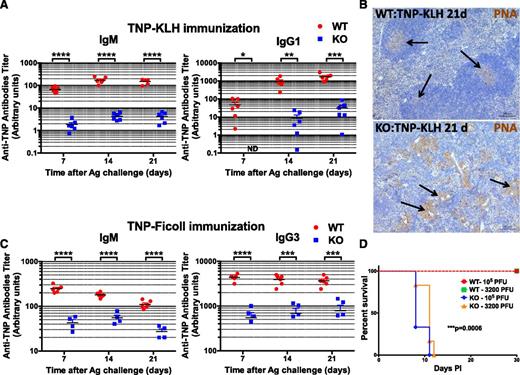
To further explore the consequences of miR-142 deletion on humoral immunity, we immunized mice with a T-cell–dependent (TD) antigen (2,4,6-trinitrophenyl conjugated to keyhole limpet hemocyanin [TNP-KLH]) and analyzed production of antigen-specific antibodies postimmunization. We found that in contrast to a robust immune response to the TNP-KLH challenge in the control mice, the antibody response in the KO mice was largely blunted (Figure 5A). Staining of spleen sections from TNP-KLH immunized mice with peanut agglutinin (PNA) revealed a lack of germinal centers in the KO organs and abnormal accumulation of PNA+ myeloid cells in the tissue (Figure 5B). Challenge of miR-142−/− mice with a T-cell–independent (TI) antigen (TNP-Ficoll) also resulted in a largely unproductive humoral immune response (Figure 5C). Since TI antigens typically do not require T cell help for B cell activation, our findings strongly imply that aberrant antibody production in the miR-142−/− mice is the result of a B-cell–specific defect. Finally, to examine the role of miR-142 in cellular immunity, we infected the KO mice with herpes simplex virus (HSV-1) and monitored their survival over time. C57BL/6 mice are normally resistant to ocular HSV-1 infection and depend on a robust T cell response to the virus for protection.19-22 However, miR-142−/− mice were highly susceptible to infection with HSV-1 and died of virus-induced encephalitis within 11 days (Figure 5D). Lowering the titer of HSV-1 by ∼30-fold did not improve the survival of miR-142−/− mice. Together, our results suggest that miR-142−/− mice develop a combined immunodeficiency and as a result are unable to mount productive immune responses to both soluble and viral antigens.
miR-142 KO mice are immunodeficient. (A-C) Defective humoral immune responses in miR-142−/− mice. (A) Analysis of anti-TNP antibody production (IgM, left; IgG1, right) in 8-week-old female WT (n = 6) and KO (n = 6) mice challenged with T-cell–dependent antigen (TNP-KLH, 100 μg in complete Freund adjuvant) by serum Ig ELISA. Peripheral blood was collected 7, 14, and 21 days postimmunization. (B) Immunohistochemical analysis of GC formation in spleen sections from WT (top) and KO (bottom) mice challenged with TNP-KLH. Spleens were collected 21 days postimmunization and stained with PNA (brown stain). Note normal GC formation (marked by arrows) in WT spleen, whereas KO tissue lacks GCs and instead displays multiple PNA+ myeloid cell clusters (marked by arrows). (C) Analysis of anti-TNP antibody production (IgM, left; IgG3, right) in 6-week-old female WT (n = 6) and KO (n = 4) mice challenged with TI antigen (TNP-Ficoll, 100 μg in phosphate-buffered saline) by serum Ig ELISA. Peripheral blood was collected 7, 14 and 21 days post immunization. (D) miR-142−/− mice are highly susceptible to infection with HSV-1 virus. Kaplan-Meier survival curves of WT (n = 6) and KO (n = 6) mice upon infection with either low (3200 plaque-forming units) or high (105 plaque-forming units) dose of HSV-1 virus. P values for the comparison of WT and KO curves were identical for both HSV-1 doses and were calculated using a log-rank (Mantel-Cox) test. Results are shown as means ± SD. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. Ag, antigen; ND, not detected.
miR-142 KO mice are immunodeficient. (A-C) Defective humoral immune responses in miR-142−/− mice. (A) Analysis of anti-TNP antibody production (IgM, left; IgG1, right) in 8-week-old female WT (n = 6) and KO (n = 6) mice challenged with T-cell–dependent antigen (TNP-KLH, 100 μg in complete Freund adjuvant) by serum Ig ELISA. Peripheral blood was collected 7, 14, and 21 days postimmunization. (B) Immunohistochemical analysis of GC formation in spleen sections from WT (top) and KO (bottom) mice challenged with TNP-KLH. Spleens were collected 21 days postimmunization and stained with PNA (brown stain). Note normal GC formation (marked by arrows) in WT spleen, whereas KO tissue lacks GCs and instead displays multiple PNA+ myeloid cell clusters (marked by arrows). (C) Analysis of anti-TNP antibody production (IgM, left; IgG3, right) in 6-week-old female WT (n = 6) and KO (n = 4) mice challenged with TI antigen (TNP-Ficoll, 100 μg in phosphate-buffered saline) by serum Ig ELISA. Peripheral blood was collected 7, 14 and 21 days post immunization. (D) miR-142−/− mice are highly susceptible to infection with HSV-1 virus. Kaplan-Meier survival curves of WT (n = 6) and KO (n = 6) mice upon infection with either low (3200 plaque-forming units) or high (105 plaque-forming units) dose of HSV-1 virus. P values for the comparison of WT and KO curves were identical for both HSV-1 doses and were calculated using a log-rank (Mantel-Cox) test. Results are shown as means ± SD. P values were calculated using Student t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. Ag, antigen; ND, not detected.
Deletion of miR-142 results in global derepression of miR-142-3p targets
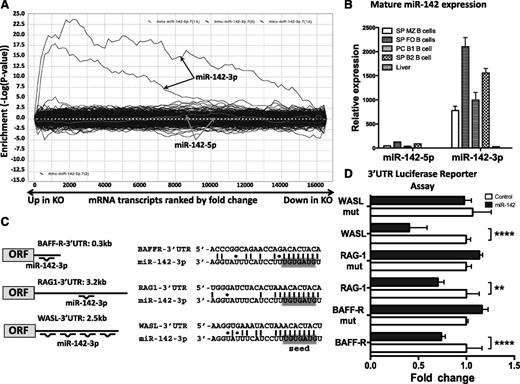
We investigated further how miR-142 deletion can lead to the multiple B cell defects observed in our mutant mice. To this end, we performed expression profiling of purified CD19+ WT and KO splenic B cells using DNA microarrays. We found that deletion of miR-142 affects expression of 624 genes (fold change > 1.2 or < −1.2; P < .01 with FDR); 396 genes were upregulated, whereas 228 were repressed in miR-142−/− B cells. Analysis of differentially expressed genes by the SylArray software algorithm23 revealed global derepression of miR-142-3p targets, but not miR-142-5p targets, in miR-142−/− B cells (Figure 6A). SylArray computes enrichment scores for miRNA seed sequences in the 3′ untranslated regions (UTRs) of differentially expressed genes; thus, it can reveal the impact of a specific miRNA gene deletion on the cell transcriptome. Our findings suggest that changes in B-cell physiology in miR-142−/− mice are most likely the result of abrogation of miR-142-3p expression, whereas miR-142-5p has little contribution to the control of gene expression. This conclusion is in agreement with the significantly lower expression of mature miR-142-5p compared with mature miR-142-3p levels in several B-cell subsets (Figure 6B).
Global derepression of miR-142-3p, but not miR-142-5p, targets in miR-142−/− B cells. (A) Analysis of the effect of miR-142 deletion on global gene expression. SylArray plot analysis for the seed complementary region (SCR) words corresponding to 2 7-mer seeds of miR-142-3p (marked by black arrows) and 2 7-mer seeds of miR-142-5p (marked by gray arrows). Log10-transformed enrichment P values for each SCR word, relative to P values of all other words, are plotted on the y-axis, against the ranked gene list on the x-axis (left, the most upregulated in KO vs WT genes; right, the most downregulated in KO vs WT genes). (B) Analysis of mature miR-142-5p and miR-142-3p expression in C57BL/6 B-cell subsets by quantitative reverse-transcription polymerase chain reaction. B-cell fractions were sorted by FACS from spleen (SP) and peritoneal cavity (PC). Expression level of miR-142-5p in liver was arbitrarily set to 1. SnoRNA234 levels were used for normalization. (C) Diagrams (left) and sequence alignments (right) of miR-142-3p putative binding sites in the 3′ UTRs of BAFF-R, RAG1, and WASL predicted by the TargetScan algorithm. (D) Validation of BAFF-R, RAG1, and WASL as direct miR-142-3p targets by 3′ UTR luciferase reporter assay. Relative expression of WT and mutated BAFF-R, RAG1, and WASL 3′ UTR reporter constructs upon cotransfection with either miR-142 expressing plasmid or empty control vector. Results are shown as means ± SD and are representative of at least 2 independent experiments. P values were calculated using 2-way analysis of variance. **P ≤ .01; ***P ≤ .001; ****P ≤ .0001.
Global derepression of miR-142-3p, but not miR-142-5p, targets in miR-142−/− B cells. (A) Analysis of the effect of miR-142 deletion on global gene expression. SylArray plot analysis for the seed complementary region (SCR) words corresponding to 2 7-mer seeds of miR-142-3p (marked by black arrows) and 2 7-mer seeds of miR-142-5p (marked by gray arrows). Log10-transformed enrichment P values for each SCR word, relative to P values of all other words, are plotted on the y-axis, against the ranked gene list on the x-axis (left, the most upregulated in KO vs WT genes; right, the most downregulated in KO vs WT genes). (B) Analysis of mature miR-142-5p and miR-142-3p expression in C57BL/6 B-cell subsets by quantitative reverse-transcription polymerase chain reaction. B-cell fractions were sorted by FACS from spleen (SP) and peritoneal cavity (PC). Expression level of miR-142-5p in liver was arbitrarily set to 1. SnoRNA234 levels were used for normalization. (C) Diagrams (left) and sequence alignments (right) of miR-142-3p putative binding sites in the 3′ UTRs of BAFF-R, RAG1, and WASL predicted by the TargetScan algorithm. (D) Validation of BAFF-R, RAG1, and WASL as direct miR-142-3p targets by 3′ UTR luciferase reporter assay. Relative expression of WT and mutated BAFF-R, RAG1, and WASL 3′ UTR reporter constructs upon cotransfection with either miR-142 expressing plasmid or empty control vector. Results are shown as means ± SD and are representative of at least 2 independent experiments. P values were calculated using 2-way analysis of variance. **P ≤ .01; ***P ≤ .001; ****P ≤ .0001.
Scrutiny of the list containing the most upregulated transcripts in miR-142−/− B cells revealed a number of genes with putative miR142-3p binding sites that have well-characterized roles in B-cell development and function (Table 1). In agreement with the notion that miRNAs can fine-tune cellular responses by targeting signaling nodes,24 some of the putative miR-142-3p target genes found to be derepressed in the KO B cells could be arranged into signaling networks. For example, miR-142−/− B cells display simultaneous upregulation of Wiskott-Aldrich syndrome–like (WASL), cofilin 2 (CFL2), actin filament associated protein 1 (AFAP1), destrin (DSTN), and ras homolog family member Q (RHOQ) genes, all of which function in the actin cytoskeleton remodeling pathway (Table 1). Moreover, recombination activating gene 1 (RAG1), RAG2, and terminal deoxynucleotidyltransferase (DNTT) play key roles in the generation of antigen receptor diversity through VDJ recombination, and all were found among the genes upregulated in miR-142−/− cells. The third signaling circuit that was also highly represented among the derepressed genes was the BAFF-R pathway, which includes BAFF-R, tumor necrosis factor receptor–associated factor 3 (TRAF3), and p52/p100 nuclear factor κB (NF-κB) subunit (NFKB2).
List of genes upregulated in miR-142−/− B cells that have a known role in B cell development and function
| Gene symbol . | Gene title . | Fold change KO vs WT . | P . | B-cell–related process . |
|---|---|---|---|---|
| RAG1 | Recombination activating gene 1 | 18.2635 | .0002 | VDJ recombination pathway ● |
| VPREB1 | Pre–B-lymphocyte gene 1 | 11.2706 | 1.12E−06 | Pre-BCR receptor complex ▪ |
| DNTT | Deoxynucleotidyltransferase, terminal | 4.8675 | 9.4E−05 | VDJ recombination pathway ● |
| AFAP1 | Actin filament–associated protein 1 | 3.6832 | 2.66E−05 | Actin cytoskeleton rearrangement ○ |
| IGLL1 | Immunoglobulin λ-like polypeptide 1 | 3.476 | .0005 | Pre-BCR receptor complex ▪ |
| DSTN | Destrin | 2.9908 | .0003 | Actin cytoskeleton rearrangement ○ |
| MYB | Myeloblastosis oncogene | 2.6885 | .001 | Transcription regulation |
| RHOQ | ras homolog gene family, member Q | 2.4311 | 4.67E−07 | Actin cytoskeleton rearrangement ○ |
| CD93 | CD93 antigen | 2.4244 | .0016 | Cell adhesion |
| RAG2 | Recombination activating gene 2 | 2.3119 | .0003 | VDJ recombination pathway ● |
| CFL2 | Cofilin 2, muscle | 2.2724 | 6.85E−05 | Actin cytoskeleton rearrangement ○ |
| WASL | Wiskott-Aldrich syndrome–like (human) | 1.8902 | 4.63E−05 | Actin cytoskeleton rearrangement ○ |
| TGFBR1 | TGF, β receptor I | 1.6585 | .0008 | TGF-β receptor signaling pathway |
| IL6ST | Interleukin-6 signal transducer | 1.613 | .0002 | IL-6 receptor signaling pathway |
| TNFRSF13c/BAFF-R | TNF receptor superfamily, member13C | 1.4771 | .0002 | BAFF-R signaling pathway ♦ |
| NFKB2 | Nuclear factor of κ light polypeptide gene enhancer in B cells 2, p49/pl00 | 1.3369 | .0033 | BAFF-R signaling pathway ♦ |
| TRAF3 | TNF receptor–associated factor 3 | 1.2257 | .0096 | BAFF-R signaling pathway ♦ |
| Gene symbol . | Gene title . | Fold change KO vs WT . | P . | B-cell–related process . |
|---|---|---|---|---|
| RAG1 | Recombination activating gene 1 | 18.2635 | .0002 | VDJ recombination pathway ● |
| VPREB1 | Pre–B-lymphocyte gene 1 | 11.2706 | 1.12E−06 | Pre-BCR receptor complex ▪ |
| DNTT | Deoxynucleotidyltransferase, terminal | 4.8675 | 9.4E−05 | VDJ recombination pathway ● |
| AFAP1 | Actin filament–associated protein 1 | 3.6832 | 2.66E−05 | Actin cytoskeleton rearrangement ○ |
| IGLL1 | Immunoglobulin λ-like polypeptide 1 | 3.476 | .0005 | Pre-BCR receptor complex ▪ |
| DSTN | Destrin | 2.9908 | .0003 | Actin cytoskeleton rearrangement ○ |
| MYB | Myeloblastosis oncogene | 2.6885 | .001 | Transcription regulation |
| RHOQ | ras homolog gene family, member Q | 2.4311 | 4.67E−07 | Actin cytoskeleton rearrangement ○ |
| CD93 | CD93 antigen | 2.4244 | .0016 | Cell adhesion |
| RAG2 | Recombination activating gene 2 | 2.3119 | .0003 | VDJ recombination pathway ● |
| CFL2 | Cofilin 2, muscle | 2.2724 | 6.85E−05 | Actin cytoskeleton rearrangement ○ |
| WASL | Wiskott-Aldrich syndrome–like (human) | 1.8902 | 4.63E−05 | Actin cytoskeleton rearrangement ○ |
| TGFBR1 | TGF, β receptor I | 1.6585 | .0008 | TGF-β receptor signaling pathway |
| IL6ST | Interleukin-6 signal transducer | 1.613 | .0002 | IL-6 receptor signaling pathway |
| TNFRSF13c/BAFF-R | TNF receptor superfamily, member13C | 1.4771 | .0002 | BAFF-R signaling pathway ♦ |
| NFKB2 | Nuclear factor of κ light polypeptide gene enhancer in B cells 2, p49/pl00 | 1.3369 | .0033 | BAFF-R signaling pathway ♦ |
| TRAF3 | TNF receptor–associated factor 3 | 1.2257 | .0096 | BAFF-R signaling pathway ♦ |
Analysis of gene expression in WT (n = 3) and KO (n = 3) CD19+ B cells was performed using DNA microarrays. miR-142-3p putative target genes are indicated in bold.
TGF, transforming growth factor; TNF, tumor necrosis factor.
We employed 3′ UTR luciferase reporter assays to validate BAFF-R, RAG1 and WASL as miR-142-3p direct targets. According to TargetScan25,26 analysis, the 3′ UTR of mouse BAFF-R gene contains 1 conserved miR-142-3p binding site, whereas the 3′ UTR of mouse WASL has 2 conserved and 2 nonconserved sites (Figure 6C). The single binding site in the mouse RAG1 3′ UTR is not evolutionary conserved (Figure 6C) but can still be bound by miR-142-3p in other species if nonstandard Watson-Crick base pairing is taken into account (supplemental Figure 4). We cloned the 3′ UTRs of WASL, RAG1, and BAFF-R genes downstream of a luciferase reporter and cotransfected them together with a miR-142 expression vector in 293T cells. Expression of all 3 reporter genes significantly decreased in the presence of miR-142, but not after transfection with a control vector (Figure 6D). Introduction of mutations into the predicted binding sites in the 3′ UTRs of BAFF-R, RAG1, and WASL interfered with the ability of miR-142 to downregulate reporter expression, suggesting that miR-142-3p binds directly to these sites (Figure 6D).
Derepression of BAFF-R in miR-142−/− B cells results in constitutive activation of downstream signaling and robust proliferation in vitro
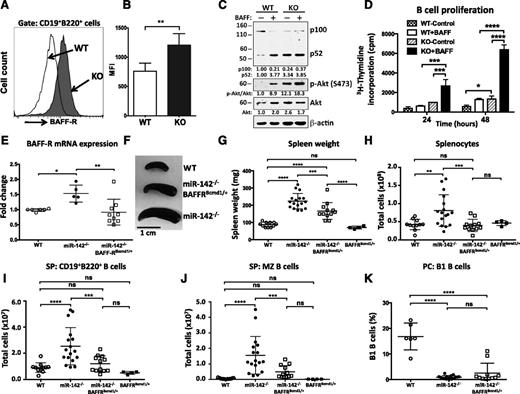
Because BAFF-R functions as a direct target of miR-142-3p, we decided to test the notion that dysregulation of this gene contributes to the development of B-cell defects in miR-142−/− mice. We focused on BAFF-R because of its well-established role in the control of peripheral B-cell growth and survival.27,28 BAFF-R is a member of the tumor necrosis factor receptor superfamily that exerts its functions by triggering activation of the noncanonical NF-κB signaling pathway29 and the Akt kinase cascade.30 Analysis of BAFF-R expression on the surface of miR-142−/− B cells by flow cytometry revealed a significant induction of BAFF-R levels (Figure 7A-B). Stimulation of purified WT B cells with BAFF ligand in vitro resulted in processing of the p100/NFKB2 precursor and subsequent accumulation of the p52 NF-κB subunit (Figure 7C). In contrast, miR-142−/− B cells displayed constitutive activation of the alternative NF-κB pathway (Figure 7C). Moreover, activation of the prosurvival Akt pathway was also altered in miR-142−/− B cells. Phosphorylated Akt protein was readily detectable in unstimulated cells, and the phosphorylation level was further elevated in response to BAFF ligand. To test how these signaling changes affect miR-142−/− B-cell functions, we examined proliferative responses of WT and KO B cells. We found that unstimulated miR-142−/− B cells proliferated at a rate similar to BAFF-activated WT cells, whereas stimulation with BAFF ligand strongly augmented their proliferative capacity (Figure 7D).
miR-142 controls B-cell homeostasis by targeting BAFF-R. (A) Upregulation of BAFF-R expression on the cell surface of miR-142−/− B cells. FACS analysis of CD19+B220+ B cells from WT (open histogram) and KO (shaded histogram) animals with anti-CD268 (BAFF-R) antibodies. (B) Mean fluorescence intensity (MFI) of BAFF-R expression in B cells from WT (n = 8) and KO (n = 6) animals. (C) Activation of noncanonical NF-κB and Akt pathways in miR-142−/− cells. Western blot analysis of p100/NFKB2 processing and Akt (S473) phosphorylation in purified CD19+ B cells isolated from WT and KO animals. Cells were stimulated with human BAFF ligand (500 ng/mL) for 15 hours. β-Actin and pan-Akt signals were used as loading controls. (D) Analysis of B-cell proliferation by 3H-thymidine incorporation assay. Purified WT (n = 2) and KO (n = 2) B cells were stimulated with human BAFF ligand (500 ng/mL) for 24 and 48 hours. (E-K) Lowering of the BAFF-R gene dose rescues the B-cell expansion defect in miR-142−/− mice. (E) Analysis of BAFF-R expression in WT (n = 6), miR-142−/− (n = 5), and miR-142−/−BAFF-R+/BCMD1 (n = 9) splenocytes by quantitative reverse-transcription polymerase chain reaction. Expression level of BAFF-R allele in WT mouse was arbitrarily set to 1. Mouse β-actin levels were used for normalization. (F-H) Rescue of the splenomegaly phenotype in miR-142−/−BAFF-R+/BCMD1 mice. (F) Representative image of spleens from WT, miR-142−/− and miR-142−/−BAFF-R+/BCMD1 mice. Spleen weights (G) and total splenocyte counts (H) in WT (n = 9), miR142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) animals. (I) Total CD19+B220+ B-cell counts in WT (n = 9), miR-142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) spleens. (J) Normal number of MZ B cells in miR-142−/−BAFF-R+/BCMD1 mice. Total number of MZ B cells in WT (n = 9), miR-142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) spleens. (K) Lack of rescue of the B1 B-cell defect in miR-142−/−BAFF-R+/BCMD1 mice. Total number of B1 B cells in the peritoneal cavities of WT (n = 6), miR-142 KO (n = 16), and miR-142−/−BAFF-R+/BCMD1 (n = 11) mice. Results are plotted as means ± SD. P values were calculated using Student t test or 2-way analysis of. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. ns, not significant.
miR-142 controls B-cell homeostasis by targeting BAFF-R. (A) Upregulation of BAFF-R expression on the cell surface of miR-142−/− B cells. FACS analysis of CD19+B220+ B cells from WT (open histogram) and KO (shaded histogram) animals with anti-CD268 (BAFF-R) antibodies. (B) Mean fluorescence intensity (MFI) of BAFF-R expression in B cells from WT (n = 8) and KO (n = 6) animals. (C) Activation of noncanonical NF-κB and Akt pathways in miR-142−/− cells. Western blot analysis of p100/NFKB2 processing and Akt (S473) phosphorylation in purified CD19+ B cells isolated from WT and KO animals. Cells were stimulated with human BAFF ligand (500 ng/mL) for 15 hours. β-Actin and pan-Akt signals were used as loading controls. (D) Analysis of B-cell proliferation by 3H-thymidine incorporation assay. Purified WT (n = 2) and KO (n = 2) B cells were stimulated with human BAFF ligand (500 ng/mL) for 24 and 48 hours. (E-K) Lowering of the BAFF-R gene dose rescues the B-cell expansion defect in miR-142−/− mice. (E) Analysis of BAFF-R expression in WT (n = 6), miR-142−/− (n = 5), and miR-142−/−BAFF-R+/BCMD1 (n = 9) splenocytes by quantitative reverse-transcription polymerase chain reaction. Expression level of BAFF-R allele in WT mouse was arbitrarily set to 1. Mouse β-actin levels were used for normalization. (F-H) Rescue of the splenomegaly phenotype in miR-142−/−BAFF-R+/BCMD1 mice. (F) Representative image of spleens from WT, miR-142−/− and miR-142−/−BAFF-R+/BCMD1 mice. Spleen weights (G) and total splenocyte counts (H) in WT (n = 9), miR142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) animals. (I) Total CD19+B220+ B-cell counts in WT (n = 9), miR-142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) spleens. (J) Normal number of MZ B cells in miR-142−/−BAFF-R+/BCMD1 mice. Total number of MZ B cells in WT (n = 9), miR-142−/− (n = 17), miR-142−/−BAFF-R+/BCMD1 (n = 12), and BAFF-R+/BCMD1 (n = 4) spleens. (K) Lack of rescue of the B1 B-cell defect in miR-142−/−BAFF-R+/BCMD1 mice. Total number of B1 B cells in the peritoneal cavities of WT (n = 6), miR-142 KO (n = 16), and miR-142−/−BAFF-R+/BCMD1 (n = 11) mice. Results are plotted as means ± SD. P values were calculated using Student t test or 2-way analysis of. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. ns, not significant.
Lowering the BAFF-R gene dosage rescues the B-cell expansion defect in miR-142−/− mice
Because miR-142−/− B cells express more BAFF-R, we reasoned that lowering the BAFF-R gene dose would allow us to dissect the functional role of a miR-142-3p-BAFF-R interaction in vivo. To this end, we crossed miR-142−/− mice with the A/WySnJ strain that carries a naturally occurring B-cell maturation defect-1 (BCMD-1) mutation. BCMD-1 is a loss-of-function mutation that results from a retrotransposon insertion into exon 3 of the BAFF-R gene, encoding the cytoplasmic domain of BAFF-R.31,32 The resulting miR-142−/−BAFF-R+/BCMD1 mice carry only 1 functional BAFF-R allele and show, as expected, a decrease in the expression of BAFF-R mRNA in B cells down to WT levels (Figure 7E). Gross morphologic analysis of miR-142−/−BAFF-R+/BCMD1 mice revealed partial rescue of the splenomegaly defect (Figure 7F-G), whereas the total number of splenocytes was significantly reduced (Figure 7H). Moreover, lowering the BAFF-R gene dose also decreased the number of CD19+B220+ B cells in miR-142−/− mice and subsequently rescued the B-cell expansion defect (Figure 7I and supplemental Figure 5A). miR-142−/−BAFF-R+/BCMD1 spleens displayed normal levels of MZ B cells (Figure 7J), whereas the number of FO B cells dropped slightly in comparison with WT mice (supplemental Figure 5B). We did not observe phenotypic rescue of the B1 B-cell or hypogammaglobulinemia defects in miR-142−/−BAFF-R+/BCMD1 mice (Figure 7K and data not shown). Taken together, our findings suggest that miR-142-3p-BAFF-R regulatory axis controls the size of the B2 B-cell compartment, whereas other miR-142-3p targets are probably mediating its effects on humoral immunity and B1 B-cell differentiation.
Discussion
With a relatively simple mechanism of target recognition, miRNAs have the potential for high evolutionary plasticity, making them ideal regulators of innate and adaptive immunity in organisms continually presented with new pathogenic challenges.33 The preferential and abundant expression of miR-142 in hematopoietic tissue led us to test the hypothesis that it controls certain aspects of immune homeostasis and function.
Our findings in miR-142−/− mice implicate this miRNA as a critical regulator of lymphopoiesis. The KO mice display an enlarged splenic B-cell compartment, mostly due to abnormal expansion of MZ-like B cells, as well as contraction of T- and B1 B-cell populations in the periphery. miR-142 plays a cell-intrinsic role in the regulation of lymphocyte differentiation because chimeric mice reconstituted with miR-142−/− BM develop the same B- and T-cell defects as the miR-142 KO animals. Although the role of miR-142 in lymphocyte development was not a subject of intense research in the past, our observations agree well with prior studies. For example, ectopic expression of miR-142 in BM progenitor cells was shown to hinder B-cell development in vitro.7 Furthermore, conditional deletion of Dicer in B cells resulted in preferential expansion of MZ B cells,3 whereas abrogation of Dicer expression in CD4+ cells has been shown to reduce the cellularity of the peripheral T-cell compartment.5 These results, taken together with the abundance of miR-142 expression in immune cells, make it a prime candidate for a miRNA gene whose deletion is driving the above-mentioned immune defects in the Dicer-deficient mice.
Our results indicate that miR-142 is not only required for normal development of lymphocytes but also indispensable for the optimal function of mature B and T cells. We report herein that miR-142−/− mice develop a combined immunodeficiency with defects in both humoral and cellular immunity. Challenges of the miR-142−/− mice with TD and TI antigens failed to yield a robust antibody response. At the same time, miR-142−/− mice are highly susceptible to infection with HSV-1 virus, an infection model that largely depends on functional CD4+ and CD8+ T cells for protection.20,22,34 Our findings suggest that miR-142−/− mice display a genuine defect in mature B-cell function based on the following observations: (1) failure of the miR-142−/− mice to mount an effective antibody response to challenge with TI antigen (an immune reaction that arguably depends only on B cells) and (2) defective generation of short-term plasma cells from splenic B cells in vitro. This conclusion is also indirectly supported by the similarity of defects in miR-142−/− mice and mice with a specific deletion of Dicer in antigen-experienced B cells, which display impaired generation of GCs, plasma cells, and high-affinity antibodies in response to antigen challenges.4
B-cell defects in miR-142−/− mice seem to be driven mainly by the loss of miR-142-3p activity, whereas the absence of miR-142-5p expression has little impact. Two pieces of evidence support this notion, including the global derepression of miR-142-3p and not miR-142-5p target genes in miR-142−/− B cells, as well as the higher abundance of miR-142-3p compared with miR-142-5p in immune cells. Our conclusion is in agreement with observations by others that miR-142-3p and not miR-142-5p is responsible for the regulation of immune cell function in mice12,13,35 and zebrafish.9-11
Growing evidence suggests that miRNAs tightly control cellular responses and organismal homeostasis by targeting signaling nodes in a variety of physiological networks. In agreement with this notion, our study shows that miR-142-3p targets several signaling pathways that play well-established roles in B-cell differentiation and function, including the VDJ recombination complex, actin cytoskeleton rearrangement networks, and BAFF-R signaling cascade. We became particularly interested in the latter pathway because of its proven role in B-cell homeostasis. We provided genetic evidence that the miR-142-3p-BAFF-R axis controls the size of the B2 B-cell compartment, especially the MZ B-cell subset. Loss of miR-142-3p expression elevates the level of BAFF-R on the surface of B cells and results in constitutive activation of the downstream signaling. Consequently, miR-142−/− mice display dramatic changes in B-cell homeostasis, which to a large degree resemble defects observed in BAFF transgenic mice, including splenomegaly, expansion of the B-cell compartment, and increases in MZ B cells.36 However, not all B-cell phenotypes, such as the B1 B-cell defect and hypogammaglobulinemia, were rescued in miR-142−/− mice by lowering the BAFF-R gene dose. These results imply separation of function at the level of miR-142-3p targets and, therefore, highlight the relevance and need for future analysis of other targets of this miRNA in B-cell differentiation and function.
Although our work reveals novel insights into miR-142 function, one clinical implication of this study is a rational possibility that mutations in miR-142 gene are behind compromised immunity in patients with immunodeficiencies. Primary immunodeficiency diseases are monogenic disorders that are usually manifested by a wide range of clinical symptoms including persistent infections, autoimmunity, lymphoproliferation, and cancer.37 Intriguingly, miR-142 mutations are found in ∼20% of diffuse large B-cell lymphoma patients.38 Experimental testing of our hypothesis that miR-142 lesions are important etiologic factors for primary immunodeficiency diseases could potentially pave the way for the development of novel diagnostic and pharmacologic strategies to restore immune homeostasis in these patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sophia Loera for her help with the processing and analysis of histological samples and members of both the Boldin and Bailis laboratories for helpful discussions and suggestions.
Authorship
Contribution: N.J.K., W.-L.W., E.Y.R., and M.P.B. designed the study, developed methodology, collected and analyzed data, and wrote the manuscript; B.K. and C.-C.C. assisted with collection and analysis of data from mixed BM chimeras; C.R. and E.M.C. performed and analyzed data from HSV-1 infection experiments; K.D.T. and N.C. assisted with collection and analysis of expression profiling data; and S.L.V. performed histopathological analysis of tissue sections.
Conflict-of-interest disclosure: N.C. is a shareholder of Regulus Therapeutics, a biotech company developing miRNA-based drugs. The remaining authors declare no competing financial interests.
The current affiliation for K.D.T. is EMD Millipore, Temecula, CA.
Correspondence: Mark P. Boldin, Department of Molecular and Cellular Biology, Beckman Research Institute, City of Hope, 1500 East Duarte Rd, Duarte, CA 91010-3000; e-mail: mboldin@coh.org.
References
Author notes
N.J.K. and W.-L.W. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal