Key Points
C3 mutations in aHUS commonly result in impaired complement regulation, C3 consumption, and a poor renal outcome.
C3 mutations tend to cluster at the protein surface and facilitate mapping of putative binding sites for the regulatory proteins.
Abstract
The pathogenesis of atypical hemolytic uremic syndrome (aHUS) is strongly linked to dysregulation of the alternative pathway of the complement system. Mutations in complement genes have been identified in about two-thirds of cases, with 5% to 15% being in C3. In this study, 23 aHUS-associated genetic changes in C3 were characterized relative to their interaction with the control proteins factor H (FH), membrane cofactor protein (MCP; CD46), and complement receptor 1 (CR1; CD35). In surface plasmon resonance experiments, 17 mutant recombinant proteins demonstrated a defect in binding to FH and/or MCP, whereas 2 demonstrated reduced binding to CR1. In the majority of cases, decreased binding affinity translated to a decrease in proteolytic inactivation (known as cofactor activity) of C3b via FH and MCP. These results were used to map the putative binding regions of C3b involved in the interaction with MCP and CR1 and interrogated relative to known FH binding sites. Seventy-six percent of patients with C3 mutations had low C3 levels that correlated with disease severity. This study expands our knowledge of the functional consequences of aHUS-associated C3 mutations relative to the interaction of C3 with complement regulatory proteins mediating cofactor activity.
Introduction
The complement system is an immune surveillance and effector system operating in the blood and on the cell surface. It plays a key role in defense against pathogens and in maintenance of host homeostasis.1 Genetic or acquired abnormalities in complement proteins are associated with a variety of infectious, inflammatory, autoimmune, and thrombosis-associated pathologies. Atypical hemolytic uremic syndrome (aHUS) is a rare renal thrombotic microangiopathy characterized by a mechanical hemolytic anemia, a consumptive thrombocytopenia, and acute renal failure. The renal disease is caused by formation of microthrombi in the small blood vessels of glomeruli. aHUS is a prototypical disease induced by overactivation of the alternative pathway of the complement system (Figure 1A). It is associated with a mutation in the complement regulators factor H (FH), factor I (FI), or membrane cofactor protein (MCP, CD46), as well as the activating components C3 or factor B (FB) of the central enzymatic complex of the complement alternative pathway, the C3 convertase.2
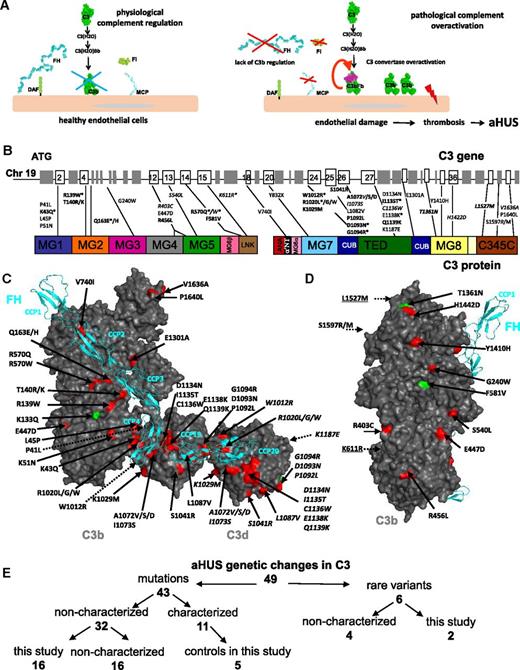
Role of complement in aHUS and localization of the aHUS mutations in C3. (A) Role of complement alternative pathway in the physiopathology of aHUS. The alternative complement pathway is continuously activated by the spontaneous transformation of the biologically inactive central complement component C3 into a biologically active form C3(H2O) to serve as a first line of defense against pathogens. C3(H2O) interacts with factor B to form the fluid phase complex C3(H2O)Bb, which has a C3 convertase activity, ie, it is able to cleave native C3 into biologically active fragments C3a (with proinflammatory activity) and C3b (which binds covalently to the cell surface). On the surface of pathogens, the complement cascade proceeds, but on healthy host cells (including the endothelial cells represented here) deposited C3b is rapidly inactivated by regulatory molecules, including MCP, FH, and FI. In case of FB binding and C3 convertase formation, it is dissociated by FH and DAF. These regulatory proteins protect healthy cells from complement overactivation and thereby prevent excessive host tissue damage. In case of mutations of the complement regulators FH, MCP, or FI, complement regulation may become inefficient. When mutations in the components of the C3 convertase (C3 and FB) are present, they may induce a formation of an overactive C3 convertase or a convertase, which is resistant to regulation. In both cases, the complement cascade is be activated on the glomerular endothelial cell surface, leading to endothelial damage, thrombosis, erythrocyte lysis–aHUS. (B) Mapping of the aHUS mutations on the C3 gene and protein. The individual exons and protein domains are indicated. Genetic changes indicated in bold are functionally characterized in this study. Rare variants are indicated in italics. Mutations marked with * are recurrent, being found in 2 or more unrelated patients from different cohorts. (C) Mapping of the aHUS mutations on the surface of C3b and C3d (in gray) in a complex with FH domains CCP1-4 and CCP19-20 (in cyan). Mutations are colored red and rare polymorphisms are green. The genetic changes characterized here are given in bold. The model is reconstructed using the structures of C3b-FH 1-48 and C3d-FH 19-20.6 (D) Side view of the C3b molecule with mapped aHUS mutations. This surface holds the binding site of the substrate molecule C3, which enters into the convertase to be cleaved. Not visible on C and D are 2 polymorphisms (indicated with dashed arrows) that are located on the surface, opposite to the FH binding site. Further, one mutation (S1597R/M) is buried in the interior of the protein, near the surface and opposite the FH binding site (indicated also with a dashed arrow). The numbering of the mutated residues is according to the mature protein sequence, lacking the 22-amino-acid signal peptide. (E) Summary of the genetic changes in C3 found in aHUS and characterized in this study.
Role of complement in aHUS and localization of the aHUS mutations in C3. (A) Role of complement alternative pathway in the physiopathology of aHUS. The alternative complement pathway is continuously activated by the spontaneous transformation of the biologically inactive central complement component C3 into a biologically active form C3(H2O) to serve as a first line of defense against pathogens. C3(H2O) interacts with factor B to form the fluid phase complex C3(H2O)Bb, which has a C3 convertase activity, ie, it is able to cleave native C3 into biologically active fragments C3a (with proinflammatory activity) and C3b (which binds covalently to the cell surface). On the surface of pathogens, the complement cascade proceeds, but on healthy host cells (including the endothelial cells represented here) deposited C3b is rapidly inactivated by regulatory molecules, including MCP, FH, and FI. In case of FB binding and C3 convertase formation, it is dissociated by FH and DAF. These regulatory proteins protect healthy cells from complement overactivation and thereby prevent excessive host tissue damage. In case of mutations of the complement regulators FH, MCP, or FI, complement regulation may become inefficient. When mutations in the components of the C3 convertase (C3 and FB) are present, they may induce a formation of an overactive C3 convertase or a convertase, which is resistant to regulation. In both cases, the complement cascade is be activated on the glomerular endothelial cell surface, leading to endothelial damage, thrombosis, erythrocyte lysis–aHUS. (B) Mapping of the aHUS mutations on the C3 gene and protein. The individual exons and protein domains are indicated. Genetic changes indicated in bold are functionally characterized in this study. Rare variants are indicated in italics. Mutations marked with * are recurrent, being found in 2 or more unrelated patients from different cohorts. (C) Mapping of the aHUS mutations on the surface of C3b and C3d (in gray) in a complex with FH domains CCP1-4 and CCP19-20 (in cyan). Mutations are colored red and rare polymorphisms are green. The genetic changes characterized here are given in bold. The model is reconstructed using the structures of C3b-FH 1-48 and C3d-FH 19-20.6 (D) Side view of the C3b molecule with mapped aHUS mutations. This surface holds the binding site of the substrate molecule C3, which enters into the convertase to be cleaved. Not visible on C and D are 2 polymorphisms (indicated with dashed arrows) that are located on the surface, opposite to the FH binding site. Further, one mutation (S1597R/M) is buried in the interior of the protein, near the surface and opposite the FH binding site (indicated also with a dashed arrow). The numbering of the mutated residues is according to the mature protein sequence, lacking the 22-amino-acid signal peptide. (E) Summary of the genetic changes in C3 found in aHUS and characterized in this study.
C3 is the central component of the complement system and is activated to C3b by proteolytic activity of the C3 convertases. Alternatively, C3 can turn over at a low rate (0.2-0.4%/h) in the serum to generate C3(H2O).3 C3b and C3(H2O) are considered to be functionally equivalent as both forms can bind to FB to form a C3 convertase and both are inactivated by regulatory proteins. This is in contrast to native C3, which does not interact with any of these ligands.
The plasma proteins FH and FI ensure complement control in plasma, whereas MCP, decay accelerating factor (DAF, CD55), and complement receptor 1 (CR1; CD35) are membrane proteins that prevent excessive C3b deposition and membrane injury. FH, DAF, and CR1 accelerate the dissociation of the AP C3 convertase, whereas FH, MCP, and CR1 have cofactor activity for C3b; ie, its cleavage and inactivation mediated by the serine protease FI. Although the mechanism of action of these regulators is well understood, questions remain regarding their binding sites on C3b and the relative importance of each regulator in inhibiting complement activation at different tissue sites. Several studies have shown that FH, MCP, and CR1 have overlapping yet distinct binding sites,4,5 but structural data are available only for FH.6-8 Also, it is unclear why aHUS is associated with mutations in MCP and FH but not DAF or CR1 because all 4 proteins regulate complement at the level of C3.
In the era of high-throughput genetic analysis, where multiple genetic changes can be found in every patient, it is of a critical importance to know if an identified genetic abnormality is disease relevant or just a fortuitous association. The aim of this study was to identify the functional consequences of a series of 23 genetic changes in C3 in aHUS patients and determine the likely mechanism whereby these changes predispose to complement alternative pathway dysregulation. We found that decreased ligand binding leading to deficient cofactor activity of FH and/or MCP was the main functional defect, although 2 of the mutations impacted CR1 binding. These results allowed us to stratify the importance of the complement regulators for the development of aHUS and to define distinct, but overlapping, surface areas on C3 important for binding FH, MCP, and CR1.
Materials and methods
Patients
C3 was directly sequenced in patients from the aHUS cohorts from France (n = 343), Italy (n = 300), the United Kingdom (n = 344), and the United States (n = 314). A literature search was also performed to collect data from all published cases of aHUS patients carrying C3 mutations. We considered as mutations only novel variants. Genetic changes reported in the 1000 Genomes Database (http://www.1000genomes.org/1000-genomes-browsers) or in the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) with a minor allele frequency <0.01 were considered rare variants.
Structural analysis
The crystal structures of C3,9 C3b,10 C3b-FH1-4,8 C3bBb,11 C3bB,12 and the C5 convertase model, based on the cobra venom factor/C5 complex13 and MCP,14 were used to map the mutated residues in Pymol. Protein numbering was based on the sequence of the mature protein, omitting the 22-amino-acid signal peptide.
Recombinant protein expression
Point mutations were introduced in the C3 cDNA in the pSV vector using site-directed mutagenesis (Quik-Change; Stratagene), and the resulting constructs were transiently or stably transfected in 293T cells as previouasly described.15,16 In some experiments, an MCP function-blocking monoclonal antibody (mAb; GB24) was added to the cultures to prevent MCP-dependent C3b degradation by complement.17 Three days after transfection, supernatants were concentrated, and mAb was removed with protein G agarose (Pierce). Alternatively, serum-free medium was used to prevent C3 degradation. Recombinant C3 was purified by ion exchange chromatography using diethylaminoethyl (DEAE) Sepharose (GE Healthcare) and eluted with 0.2 M NaCl.
Quantitation and characterization of C3 were performed by sandwich enzyme-linked immunosorbent assay and western blotting, using a polyclonal rabbit anti-C3 antibody (Calbiochem).15
Surface plasmon resonance
Surface plasmon resonance (SPR) analysis was performed using ProteOn XPR36 equipment (BioRad). Recombinant soluble MCP (sMCP),15 FH (Complement Technologies, Tyler, TX), FH CCP19-20,18 soluble CR1 (sCR1) (R&D Systems), or the anti-C3d mAb (Quidel, San Diego, CA) were coupled to individual channels of the GLC biosensor (BioRad). Purified C3 from 3e independent preparations was injected in duplicate on different chips to evaluate reproducibility of results. Alternatively, C3b or C3d (Complement Technologies) was injected over sCR1-coated surface in running buffer containing 25 or 150 mM NaCl. The kinetic fit was performed with ProteOn manager software using 1:1 Langmuir or 2-state analysis.
Cofactor assays
C3 preparations were incubated for 0 to 30 minutes at 37°C with factor I (5 ng in sMCP assays, 20 ng in FH assays, and 15 ng in CR1 assays; Complement Technologies) and sMCP (50 ng), FH (200 ng; Complement Technologies), or sCR1 (150 ng; Avant Immunotherapeutics) in 15 µL of buffer (10 mM Tris, pH 7.4, 150 mM NaCl).15,16 To stop the reaction, 7 µL of 3× reducing Laemmli sample buffer was added. Samples were boiled at 95°C for 5 min, electrophoresed on 10% Tris-glycine polyacrylamide gels, transferred to nitrocellulose, and blocked overnight with 5% non-fat dry milk in phosphate-buffered saline. Blots were probed with a 1:5000 dilution of goat anti-human C3 (Complement Technologies) followed by horseradish peroxidase-conjugated rabbit anti-goat IgG (Sigma, St. Louis, MO) and developed with SuperSignal substrate (Thermo Scientific).
Endothelial cell assays
Third-passage primary human umbilical vein endothelial cells (HUVECs) were used as described.16,19,20 Briefly, HUVECs, either resting or stimulated with tumor necrosis factor-α/interferon-γ, were exposed to 33% normal human sera from 3 individual donors, patient sera containing either the R139W or A1072D mutations, or a serum immuno-depleted of FH (FH-dpl). Activated HUVEsC were used to ensure stronger complement activation to test its modulation by FH.16,19 Sera were supplemented with different concentrations of purified FH (Complement Technologies). Cells were detached and probed for C3 activation fragment deposition with a mouse anti-human C3c mAb (Quidel), followed by a secondary anti-mouse IgG-PE (Beckman Coulter) and analyzed by flow cytometry.
Results
Cohorts’ analysis and review of the literature
The French, Italian, UK, and US aHUS cohorts were screened for genetic complement abnormalities, and 19, 11, 9, and 4 genetic changes in the C3 gene were identified, respectively (supplemental Tables 1 and 2 available on the Blood Web site). The C3 mutation frequency was 11.4%, 4.6%, 6.1%, and 4.5% for these 4 cohorts, respectively. These data, together with cases published in the literature, gave a total of 48 different genetic changes in C3 that have been associated with aHUS in 130 patients (supplemental Table 2). Of these patients, 41% were pediatric cases (<18 years of age) and 56% were female. Sporadic aHUS was found in 97 patients, and 33 patients were identified in 14 different pedigrees.15,16,21-33
Six of the observed genetic changes have been reported in 1000 Genomes or the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (NHLBI GO ESP) (R403C, S540L, K611R, T1361N, L1527M, and S1597R; supplemental Table 1). Fifteen of the 42 mutations were recurrent (identified in ≥2 unrelated aHUS patients from the same or different cohorts; Figure 1; supplemental Table 2). The genetic changes were not randomly distributed but rather preferentially clustered in exons 2 to 5, 12 to 15, and 24 to 27 (Figure 1B). They tended to map to the protein surface, with 27 of 48 variants colocalizing or being adjacent to FH CCP1-4 (especially CCP3-4) and CCP19-20 binding sites on C3b6-8 (Figure 1C). Among these 27, 21 mutations resulted in a loss, gain, or exchange of a charged residue. Two mutated residues were in proximity to the Bb binding site (V1636A and P1640L), one was in the binding site for the C3 convertase substrate C311 or C513 (R456L), and another was adjacent to the FI cleavage site in the CUB domain (E1301A). Seventeen variants were remote from the binding site for FH, MCP, CR1, CR2, CR3, or FB (Figure 1D).
Protein expression
To functionally characterize the C3 variants identified, we used a mammalian expression system. We were able to recombinantly express 23 of the mutant proteins identified in 54 patients from the different aHUS cohorts (33 from the French cohort, 10 from the UK cohort, 8 from the Italian cohort, and 3 from the US cohort; Figure 1B-D). Of these variants, 21 were completely novel, whereas 2 were reported in the 1000 Genomes or NHLBI GO Exome Sequencing Project (NHLBI GO ESP) databases (T1361N and L1527M; Figure 1E). Eighteen have not been studied, and 5 previously characterized variants served as controls. The other C3 variants identified were not expressed at a high enough level for study or were identified in patients after January 2014.
The recombinant proteins used for the functional characterization were in the form of C3(H2O). This was confirmed by their failure to undergo autolytic cleavage,34 lack of hemolytic activity, and ability of the recombinant C3 to interact with the regulatory proteins (data not shown). C3(H2O), despite the presence of the ANA domain (C3a), is thought to be conformationally very similar to C3b as it binds to the same ligands and is regulated in the same fashion.3,35-38 Therefore, we used these recombinant proteins as a model to characterize C3(H2O) and C3b interactions, especially in low ionic strength buffers, where the 2 structures are very similar.39
Binding to regulatory proteins
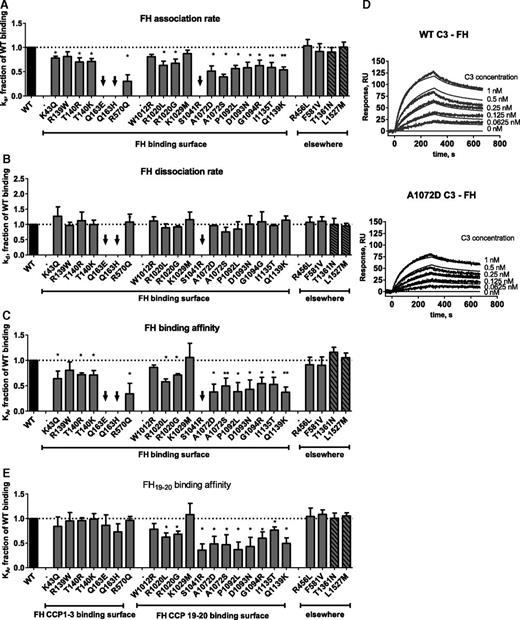
The influence of each of the 23 C3 genetic changes on binding to full-length FH, FH CCP19-20, sMCP, and sCR1 was tested by SPR (Figures 2-4; supplemental Table 3) and confirmed by enzyme-linked immunosorbent assay (data not shown). Under the experimental conditions used (ie, low ionic strength buffer), the following average kinetic parameters were obtained for the wild-type (WT) binding: FH, ka = 5.32 × 105 Ms−1, kd = 4.38 × 10−3s−1, KD = 8.2 nM; MCP, ka = 7.64 × 104 Ms−1, kd = 5.89×10−3s−1, KD = 77 nM; and CR1, ka = 7.74 × 105 Ms−1, kd = 3.24 × 10−3s−1, KD = 4.2 nM. Of note, KA = 1/KD. The results are presented as a fraction of the affinity (KA) of WT binding in the figures and are given in nanomolar in supplemental Table 3. Mutations that map to the FH binding sites, except R139W, W1012R, and K1029M, resulted in decreased binding affinity (<75% of WT binding) for full-length FH (Figure 2A-D). To gain further insight into the interaction between C3b and FH, the binding of mutant C3 to FH CCP19-20 was then tested (Figure 2E). As expected, this interaction was unaffected by mutations that localize to the binding site of CCP3, R139W, T140R/K, Q163E/H, and R570Q (Figure 1C) but was reduced in mutations that map to the binding sites for CCP196,7 and also CCP48 and CCP20.6 All but 2 mutations that alter FH binding also affected the interaction with MCP (exceptions, Q193E and Q163H; Figure 3). Two mutations in the thiol-ester domain (S1041R and P1092L) reduced binding to sCR1 (Figure 4). One mutation (R139W) affected predominantly MCP binding and function. Despite the localization of the mutations in the FH and MCP binding sites, we cannot exclude that some of them may induce a conformational change, especially in the CUB (complement C1r/C1s, Uegf, Bmp1) and TED (thioester-containing) domains in C3b and C3(H2O), which undergo large structural changes after the cleavage of C3 to C3b.
Influence of the mutations on FH binding. Kinetic analyses were performed by SPR with immobilized FH or FH CCP19-20. The running buffer was composed of 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES; pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constants for FH binding. (B) Dissociation rate constants for FH binding. (C) Affinity for FH binding. (D) Example of SPR sensorograms and kinetic fits for FH binding. (E) Affinity for FH CCP19-20 binding. The black arrows represent mutant proteins for which the binding was low and kinetic constants could not be calculated. *P < .05; **P < .005, t test.
Influence of the mutations on FH binding. Kinetic analyses were performed by SPR with immobilized FH or FH CCP19-20. The running buffer was composed of 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES; pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constants for FH binding. (B) Dissociation rate constants for FH binding. (C) Affinity for FH binding. (D) Example of SPR sensorograms and kinetic fits for FH binding. (E) Affinity for FH CCP19-20 binding. The black arrows represent mutant proteins for which the binding was low and kinetic constants could not be calculated. *P < .05; **P < .005, t test.
Influence of the mutations on MCP binding. Kinetic analyses were performed by SPR with recombinant soluble MCP on the chip. The running buffer was composed of 10 mM HEPES (pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constants for MCP binding. (B) Dissociation rate constants for MCP binding. (C) Affinity for MCP binding. (D) Example of SPR sensorograms and kinetic fits for MCP binding. (E) Model of the putative MCP binding site, obtained by molecular modeling and based on the structures of FH CCP1-48 and MCP.14 MCP CCP1-4 are indicated in cyan. Mutations are mapped in red. *P < .05; **P < .005, t test.
Influence of the mutations on MCP binding. Kinetic analyses were performed by SPR with recombinant soluble MCP on the chip. The running buffer was composed of 10 mM HEPES (pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constants for MCP binding. (B) Dissociation rate constants for MCP binding. (C) Affinity for MCP binding. (D) Example of SPR sensorograms and kinetic fits for MCP binding. (E) Model of the putative MCP binding site, obtained by molecular modeling and based on the structures of FH CCP1-48 and MCP.14 MCP CCP1-4 are indicated in cyan. Mutations are mapped in red. *P < .05; **P < .005, t test.
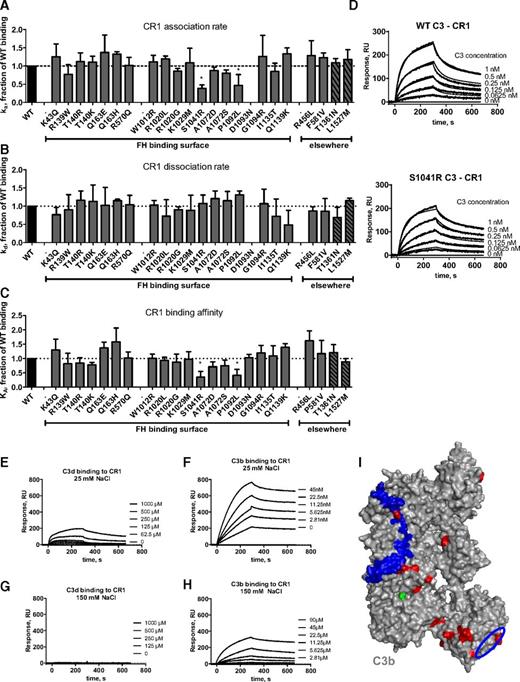
Influence of the mutations on CR1 binding. Kinetic analyses were performed by SPR with recombinant soluble CR1 immobilized on the chip. The running buffer was composed of 10 mM HEPES (pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constant for CR1 binding. (B) Dissociation rate constant for CR1 binding. (C) Affinity for CR1 binding. (D) Example of SPR sensorograms and kinetic fit for MCP binding. (E) SPR analysis of the binding of C3d to CR1 in low ionic strength buffer. (F) C3b binding to CR1 in the same buffer. (G) Lack of binding of C3d to immobilized CR1 in normal ionic strength buffer. (H) The binding of C3b to immobilized CR1 in the same buffer. (I) Location of the putative CR1 binding site on C3b, comprising residues 727 to 767 of the α′NT region5 in blue and the suggested second binding site in the TED domain in a blue circle. Mutations are mapped in red. *P < .05; **P < .005, t test.
Influence of the mutations on CR1 binding. Kinetic analyses were performed by SPR with recombinant soluble CR1 immobilized on the chip. The running buffer was composed of 10 mM HEPES (pH 7.4), 0.005% Tween-20, and 25 mM NaCl. The recombinant C3 proteins were injected for 300 seconds at 30 μL/min, followed by dissociation of 300 seconds. The chip was regenerated by injection of 0.5 M NaCl. The results are presented as a fraction of WT binding. (A) Association rate constant for CR1 binding. (B) Dissociation rate constant for CR1 binding. (C) Affinity for CR1 binding. (D) Example of SPR sensorograms and kinetic fit for MCP binding. (E) SPR analysis of the binding of C3d to CR1 in low ionic strength buffer. (F) C3b binding to CR1 in the same buffer. (G) Lack of binding of C3d to immobilized CR1 in normal ionic strength buffer. (H) The binding of C3b to immobilized CR1 in the same buffer. (I) Location of the putative CR1 binding site on C3b, comprising residues 727 to 767 of the α′NT region5 in blue and the suggested second binding site in the TED domain in a blue circle. Mutations are mapped in red. *P < .05; **P < .005, t test.
In summary, for 17 of 23 genetic changes, we observed a clear functional defect. Six variants, R456L, F581V, W1012R, K1029M, T1361N, and L1527M, did not influence FH, MCP, or CR1 binding. Of these variants, R456L and T1361N were identified in a patient who also carries the I1135T C3 mutation, which affects both FH and MCP binding; thus, for only 4 changes (F581V, W1012R, K1029M, and L1527M) were we unable to identify a functional defect. Of note, L1527M is a rare variant identified in 1000 Genomes and ESP and may be a fortuitous association and not related to the disease.
C3d binds to CR1 in low ionic strength conditions
To determine whether C3d binds to CR1, SPR experiments were performed. The binding of C3b to immobilized CR1 was readily detectable by SPR in normal ionic strength (KD ∼ 69 nM) and was enhanced at low ionic strength (KD ∼ 1.8 nM; Figure 4). We did not detect C3d binding to CR1 at physiologic ionic strength. Because this interaction is predominantly electrostatic,40 a low ionic strength was used to detect whether there was a low-affinity interaction. C3d did interact with low affinity (KD ∼ 23 µM) with CR1 at low ionic strength (buffer containing 25 mM NaCl).
Cofactor activity
To characterize cofactor function, fluid-phase assays were performed using either MCP or FH as the cofactor protein. For 14 of the 23 recombinant proteins, we obtained sufficient protein to complete the assays and demonstrated a defect in both FH- and MCP-mediated cofactor activity in 8 mutant proteins (Figure 5). Three mutant C3 proteins had largely intact FH cofactor activity but decreased MCP cofactor activity (R139W, R570Q, and R570W), and 1 mutant protein had normal MCP activity but decreased FH cofactor activity (P1092L). Two variants (R456L and F581V) had normal activity in both assays. The mutation P1092L showed both decreased CR1 binding and decreased CR1 cofactor activity (Figure 5C-D).
Functional evaluation of the C3 mutations: cofactor activity. The cofactor activity of FH or MCP toward the cleavage of the recombinant mutant C3 proteins by FI was assessed by western blot. The appearance of the α41 band indicates the cleavage. (A) Representative image for FH. (B) Representative image for MCP. (C) Representative image for CR1. (D). Example for the quantification of the western blot for CR1. (E) Two-dimensional plot representing the fraction of cofactor activity remaining relative to WT for MCP vs FH for each mutation. The dashed lines indicate the cofactor activity in case of WT, and the solid lines represent the 25% confidence limits below which the decrease was considered relevant for the disease. (F) Two-dimensional plot representing the fraction of WT binding for MCP vs FH for each mutation as shown in Figures 2 and 3. The dashed lines indicate the WT binding, and the solid lines represent the 25% confidence limits below which the decrease of the binding was considered relevant for the disease.
Functional evaluation of the C3 mutations: cofactor activity. The cofactor activity of FH or MCP toward the cleavage of the recombinant mutant C3 proteins by FI was assessed by western blot. The appearance of the α41 band indicates the cleavage. (A) Representative image for FH. (B) Representative image for MCP. (C) Representative image for CR1. (D). Example for the quantification of the western blot for CR1. (E) Two-dimensional plot representing the fraction of cofactor activity remaining relative to WT for MCP vs FH for each mutation. The dashed lines indicate the cofactor activity in case of WT, and the solid lines represent the 25% confidence limits below which the decrease was considered relevant for the disease. (F) Two-dimensional plot representing the fraction of WT binding for MCP vs FH for each mutation as shown in Figures 2 and 3. The dashed lines indicate the WT binding, and the solid lines represent the 25% confidence limits below which the decrease of the binding was considered relevant for the disease.
C3 deposition on endothelial cells
The majority of the C3 mutations (76%) had a compound functional defect, affecting binding and regulation by both FH and MCP (Figure 5F), or in the case of R139W, MCP regulation and FB binding.16 In these patients, the dissociation of the C3 convertase by FH will be inefficient. At the same time, mutant C3b will not be fully converted to iC3b on the endothelial cell surface due to decreased binding of the 2 available cofactors for FI, FH, and MCP. A secondary gain of function derived from a primary decrease in regulation would then lead to increased deposition of C3b. To determine whether increased deposition could be detected at the cell surface level, endothelial cells (HUVECs) were exposed to either WT or variant C3 serum, and C3b deposition was assessed by fluorescence-activated cell sorter (Figure 6A-B). As predicted, in the presence of C3 mutants, more C3b was detected. More purified FH was required to reduce C3 deposition on endothelial cells exposed to C3 A1072D sera, consistent with the effect of this mutation on both FH and MCP binding. In contrast, deposition of C3b carrying the R139W mutation, which affects only MCP activity, was reduced in the presence of exogenous FH (Figure 6C).
Functional evaluation of the C3 mutations: complement activation on endothelial cells. (A) Resting or (B) cytokine-activated HUVECs were incubated with different sera, and the C3 deposition was evaluated by flow cytometery. Sera from healthy donors are represented in gray (filled area under 3 different histograms, corresponding to 3 different donors); sera from aHUS patients with C3 mutations: R139W, violet; A1072D, red; or FH depleted serum as a positive control shown in blue. The black line represents a no serum control. (C) The capacity of purified FH to control the enhanced C3 deposition from FH depleted serum or patients sera on cytokines activated HUVECs was measured by flow cytometry. The percent decrease in the C3 deposition in the presence of FH compared with the serum alone is plotted vs the FH concentration used.
Functional evaluation of the C3 mutations: complement activation on endothelial cells. (A) Resting or (B) cytokine-activated HUVECs were incubated with different sera, and the C3 deposition was evaluated by flow cytometery. Sera from healthy donors are represented in gray (filled area under 3 different histograms, corresponding to 3 different donors); sera from aHUS patients with C3 mutations: R139W, violet; A1072D, red; or FH depleted serum as a positive control shown in blue. The black line represents a no serum control. (C) The capacity of purified FH to control the enhanced C3 deposition from FH depleted serum or patients sera on cytokines activated HUVECs was measured by flow cytometry. The percent decrease in the C3 deposition in the presence of FH compared with the serum alone is plotted vs the FH concentration used.
C3 concentration in plasma
Seventy-three percent of patients had low C3 levels (data available on 62 patients). Comparing patients who developed end-stage renal disease (ESRD) to those who did not (Figure 7A-B), there was a trend toward lower C3 levels in those who developed ESRD (83% vs 47%, respectively; Figure 7B).
Correlation of the C3 levels of the aHUS patients with C3 mutations with the renal outcome. (A) Data from the French cohort. C3 levels of the patients who developed ESRD up to 1 year after the onset (n = 17) were compared with the rest of the patients (n = 13). Dashed line represents the cutoff for the lower normal range for the C3 levels in the French cohort (660 mg/L). Mean ± standard deviation was 704.9 ± 326.4 for the group of no ESRD 1 year after the onset and 501.7 ± 142.3 for the ESRD group. (B) Combined data from all identified cases in the literature. C3 levels for patients with C3 mutations and with ESRD (n = 26) or without ESRD (n = 41). Lower dashed line represents the cutoff for the lower normal range for the C3 levels as measured in plasma in the French normal population (660 mg/L). Higher dashed line is the cutoff for the lower normal range for the C3 levels in the Italian cohort (830 mg/L), measured in serum. Between these 2 levels are encompassed the lower normal ranges of the other cohorts. Mean ± standard deviation was 765.8 ± 264.8 for the group of no ESRD and 604.9 ± 270.5 for the ESRD group.
Correlation of the C3 levels of the aHUS patients with C3 mutations with the renal outcome. (A) Data from the French cohort. C3 levels of the patients who developed ESRD up to 1 year after the onset (n = 17) were compared with the rest of the patients (n = 13). Dashed line represents the cutoff for the lower normal range for the C3 levels in the French cohort (660 mg/L). Mean ± standard deviation was 704.9 ± 326.4 for the group of no ESRD 1 year after the onset and 501.7 ± 142.3 for the ESRD group. (B) Combined data from all identified cases in the literature. C3 levels for patients with C3 mutations and with ESRD (n = 26) or without ESRD (n = 41). Lower dashed line represents the cutoff for the lower normal range for the C3 levels as measured in plasma in the French normal population (660 mg/L). Higher dashed line is the cutoff for the lower normal range for the C3 levels in the Italian cohort (830 mg/L), measured in serum. Between these 2 levels are encompassed the lower normal ranges of the other cohorts. Mean ± standard deviation was 765.8 ± 264.8 for the group of no ESRD and 604.9 ± 270.5 for the ESRD group.
Discussion
Here we provide detailed mapping and functional analyses of C3 mutations in patients with aHUS. The data reported here and available in the literature indicate that 48 C3 variants have been identified in 130 aHUS patients.15,16,21-32 Two additional rare single nucleotide polymorphisms, K133Q and R713W, have been seen in aHUS patients, but their frequency does not differ from that of the normal population.23 The R139W mutation was identified in 42% (14 of 33 cases) and 78% (11 of 14 cases) of aHUS patients with C3 mutations in France16 and The Netherlands,26 respectively. Eleven of 20 reported Japanese aHUS patients carry C3 mutations, with 9 patients carrying the I1135T variant.22,33 In aggregate, 15 of 42 C3 mutations have been identified in ≥2 unrelated patients around the world, highlighting the strong link between C3 mutations and the pathophysiology of aHUS.
Atypical HUS-related C3 mutations are not randomly distributed across the protein but rather cluster on the surface predominantly around FH binding sites. Although CCP20 in FH represents a hot spot for aHUS-related mutations,41 in C3, the mutations cover the entire binding area for FH, including CCP1-4 and CCP19-20. Consistent with this topographic distribution, the majority of mutations have a clear functional consequence. We identified a FH binding defect in 17 of 23 and decreased binding to MCP in 14 of 23, whereas CR1 binding was mostly unaffected. Reduced binding correlated with decreased functional cofactor activity. Apart from the direct contribution to the binding sites, most of these mutations induce electrostatic changes and may affect C3b conformation and the location of the TED domain, similarly to the common polymorphism R80G.39 The high prevalence of functionally relevant C3 mutations in aHUS is reminiscent of the findings seen in FH42-44 and MCP45 but not FB.20
C3 mutations with decreased binding to FH map to described FH-binding sites, although the structural characterization of FH binding sites on C3b and details on the organization of the FH:C3b complex are still unclear. FH binds to C3b through 2 distinct regions, CCP1-4 and CCP19-20. Based on crystal structures, 2 non-mutually exclusive possibilities for FH CCP19-20 binding to C3b and C3d have been proposed.6,7 One possibility posits that CCP19 interacts with C3d (or the TED of C3b) and CCP20 is free to bind membrane glycosaminoglycans.7,46 Alternatively, CCP19-20 binds to 2 adjacent C3d molecules, engaging the first C3d with CCP19 and the second C3d with CCP20.6,47 The CCP20 binding site, if mapped on the C3b structure, overlaps with the CCP4 binding site, indicating that CCP20 should bind to a different molecule of C3b. Because CCP4 is not present in the FH CCP19-20 miniconstruct, the results here suggest that FH CCP19-20 can indeed bind to C3b, engaging 2 adjacent molecules especially if the density of C3b and/or C3d is high.6 Such interaction will still leave free the sialic acid binding site on CCP20, allowing triple complex formation on the cell surface.46 If C3b molecules on the cell surface are scarce, CCP20 would be free to bind GAGs.7,46
In contrast to crystallographic data on the interaction of FH with C3b, information relative to MCP and CR1 and their binding to C3b is less well defined. MCP and CR1 binding sites overlap with FH binding sites, although there is no overlap between MCP and CR1 binding sites.4,5,48,49 The first CCP of MCP does not contribute to the C3b-MCP interaction.14,49,50 A C3 mutation described in a family with dense deposit disease (901ΔDG) affects 2 residues close to the FH CCP1 binding site and results in decreased FH binding and functional activity, whereas the interaction with MCP remains unaltered, suggesting that the C3 binding with FH CCP1 does not overlap MCP binding sites.51 In contrast, CR1 interacts with an area on C3 (residues 727-767, corresponding to the α′NT region of C3b) that corresponds to the FH CCP1 binding site.5,8,48 CR1 and FH also share a binding site on C3b, most likely in the area of FH CCP1-2 binding, that does not overlap with MCP because the studied mutations do not decrease CR1 binding and function. Only 2 mutations, located in the TED, reduce CR1 binding. These residues are distant from a putative CR1 binding site. This result could be explained by a large conformational change induced by the mutation or by existence of a second CR1 binding site in the TED domain. To test this, the binding of purified C3d to CR1 was evaluated and, although no binding was detected in physiological conditions, C3d bound to CR1 in low ionic strength buffer. It is therefore tempting to speculate that a second, low-affinity CR1 binding site may exist in the TED with only limited overlap with the FH CCP19-20 binding site, in agreement with previous studies.5,52 Nonetheless, as only 2 mutations demonstrated a defect in CR1 binding, the fact that CR1 is not expressed on endothelial cells53 and further that no CR1 mutations or SNPs have been reported in aHUS patients24,54 suggest that CR1 is not a major susceptibility gene for aHUS.
aHUS patients with C3 mutations develop severe disease, leading to end-stage renal failure 1 year after onset in 55% to 65% of cases.15,16,22-24 This degree of severity is reported for patients with FH mutations23,24 and also for patients with combined genetic abnormalities in which a MCP variant is combined with another mutation.55 The majority of C3 mutations primarily affect interactions with FH and MCP but also MCP and FB, as reported for R139W.16 In these patients, the dissociation of the C3 convertase by FH is inefficient, and the mutant C3b consequently has a reduced rate of conversion to iC3b on the endothelial cell surface. Complement activation is thereby enhanced, leading to endothelial damage and thrombosis. Addition of purified FH reduced C3 deposition on endothelial cells in the presence of FH-dpl or R139W containing serum in a dose-dependent fashion, reducing the C3 deposition by 50% at a concentration of 50 μg/mL. In contrast, a mutation that affects FH and MCP binding, A1072V, required much more exogenous FH (150 μg/mL) to achieve the same amount of regulation. These results point out that in patients in whom the C3 mutations affect FH binding, addition of purified FH (if such a therapeutic were available) or plasma exchange may not be efficacious.
In general, C3 consumption is an uncommon event in aHUS and is more commonly associated with quantitative FH or FI deficiency2 and with a gain-of-function FB mutation.20,53,56 Low C3 levels are frequent in C3 glomerulopathies.57 However, plasma C3 consumption is also a hallmark of the C3 mutations in aHUS, because 73% of the patients from all cohorts for whom the data are available present with low C3. This result is similar to previous reports showing 75% to 80% with low C3.23,24 This phenomenon could be explained in part by the fact that the majority of the mutations are located within or close to the FH CCP1-4 binding interface and thus perturb fluid-phase complement regulation.
Indeed, the observed C3 consumption is similar to that seen in certain type II mutations expressed but functionally altered in the N-terminal part of FH (such as R53C, A161S, or R78G23,57-59 (V. Fremeaux-Bacchi and M. Noris, unpublished data, 2014). Nevertheless, 3 genetic changes in C3, which are distant from the FH CCP1-4 binding surface and influence only FH CCP19-20 binding (such as P1092L, D1093N, and G1094R), induce C3 consumption in patients as well. For 3 patients with G1094R and the patient with P1092L, this finding may be due to the presence of a second genetic abnormality (2 in MCP, 1 in FH, and 1 in FI). However, the 2 patients with the rare variant D1093N had a low C3 without an identifiable other cause. Despite the fact that these 3 genetic changes in C3 are far from the putative MCP binding site, they demonstrate decreased MCP binding (Figure 2). P1092L decreases the binding to CR1 as well. It could be hypothesized that these mutations may not have only a direct effect on the FH binding and function but also affect the conformation of the TED domain.
Twelve of 22 patients (55%) with normal C3 levels carry mutations for which a low C3 has been reported in other patients with the same genetic abnormality from the same family or from a different family or cohort. This suggests an influence of the triggering event and additional genetic and/or environmental factors.
We sought to correlate the location of the mutations on the C3b surface with their functional consequence and clinical outcomes of the patients. Heterogeneity of the disease outcome was seen in patients carrying the same mutations. From the 12 patients with I1157T for whom clinical data were available, 4 developed a severe disease reaching ESRD, and 8 had a relapsing disease and/or achieved remission.22-24,33 A similar observation was made previously for R139W where 9 of 14 patients had ESRD and 5 of 9 had chronic kidney disease.16 These results support the notion that a genetic abnormality with a functional consequence is required but not sufficient to determine the disease occurrence and outcome. Despite this heterogeneity, aHUS patients with C3 mutations who develop ESRD have significantly lower C3 levels compared with the patients who have relapsing disease and maintain renal function. This result was observed for the patients in the French cohort, when the C3 levels were compared between the patients with rapid ESRD (before 1 year after onset) and the rest of the cohort and was confirmed with a review of all published cases in which the patients were separated by ESRD status. Therefore, the low C3 level at disease onset may be a predictor of early ESRD. Of the available information for eculizumab-treated patients,60 there were 4 published cases with a C3 mutation (R570Q,61 R570W,28 P1640L,21 and an unspecified C3 mutation30 ) and 12 known cases from this series. In all but 1 case, the treatment was successful: 10 after transplantation and 1 in whom aHUS activity subsided completely with a late recovery of renal function of the native kidney, and, in 3 cases, remission was achieved but the renal function did not recover. In the last case, the patient died of a stroke 9 days after successful kidney transplantation under therapy with eculizumab. The limited information suggests that eculizumab is a therapeutic option for this subgroup of patients despite the fact that the C3 consumption may persist after achievement of a disease remission.28
In conclusion, we demonstrated that most aHUS C3 mutations cluster in hot spot regions spanning the FH binding interface. For the majority of tested mutations, a clear functional consequence explaining the link between the genetic change and aHUS was identified. The functional characterization of C3 mutations in these patients enhances our understanding of disease pathogenesis and severity and at the molecular level helps to define the amino acid residues involved in the MCP and CR1 binding sites.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the NHLBI GO ESP and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the Women's Health Initiative Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010).
This work was supported by grants from Agence Nationale de la Recherche (ANR Genopath 2009-2012 09geno03101I), Assistance Publique-Hôpitaux de Paris (Programme Hospitalier de Recherche Clinique (AOM08198), European Union 7th Framework Programme for Research (EU FP7) grant 2012-305608 (EURenOmics), Association for Information and Research on Genetic Renal Diseases (AIRG) France, and Institut national de la santé et de la recherche médicale (to V.F.-B.). L.T.R. was a recipient of a European Molecular Biology Organization (EMBO) Long Term Fellowship ALTF 444-2007. E.V. has a fellowship from Associazione Ricerca Trapianti Fondazione Onlus (Milan, Italy). This work was also supported in part by National Institutes of Health, Allergy and Infectious Diseases grant R01-AIO41592 and National Heart, Lung, and Blood Institute grant U54-HL112303 (to J.P.A.), a grant from the UK Medical Research Council (G0701325) (to T.G.), and grants from the Academy of Finland (255922 and 259793) and Sigrid Jusélius Foundation (to T.S.J.).
Authorship
Contribution: V.F.-B., J.P.A., and L.T.R. designed and supervised the study; E.C.S., L.T.R., T.R., S.C., and C.H. performed protein expression, functionally characterized the recombinant proteins, and analyzed the data; L.T.R. performed the structural analyses and the endothelial cell experiments; V.F.-B., P.V.-M., R.J.H.S., T.M., M.N., E.V., T.G., and V.W. performed cohort analysis and sequenced C3 gene for the 4 respective cohorts; S.J. provided vital reagents; and L.T.R., E.C.S., J.P.A., and V.F.-B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: L. Roumenina, Cordeliers Research Center, INSERM UMRS 1138, Complement and Diseases Team, 15 rue de l'Ecole de Medecine, 75006 Paris, France; e-mail: lubka.roumenina@crc.jussieu.fr; V. Fremeaux-Bacchi, Cordeliers Research Center, INSERM UMRS 1138, Complement and Diseases Team, 15 rue de l'Ecole de Medecine, 75006 Paris, France; e-mail: veronique.fremeaux-bacchi@egp.aphp.fr; or J. Atkinson, Division of Rheumatology, Department of Medicine, Washington University in St Louis, St Louis, MO 63110-1093; e-mail: jatkinso@DOM.wustl.edu.
References
Author notes
E.C.S. and L.T.R. contributed equally to this work.
J.P.A. and. V.F.-B. contributed equally to this work.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal