Abstract
An intimate relationship exists between nuclear architecture and gene activity. Unraveling the fine-scale three-dimensional structure of the genome and its impact on gene regulation is a major goal of current epigenetic research, one with direct implications for understanding the molecular mechanisms underlying human phenotypic variation and disease susceptibility. In this context, the novel revolutionary genome editing technologies and emerging new ways to manipulate genome folding offer new promises for the treatment of human disorders.
Introduction
Fundamental, yet unanswered, questions in biology are how genome organization and chromosomal folding influence basic cellular processes such as transcription, how they relate to the development of disease, and whether they can be manipulated therapeutically. In mammals, gene regulatory elements are scattered throughout the genome, collectively occupying a significant fraction of the genomic noncoding DNA content. Initially, noncoding DNA (comprising ∼98% of the human genome) was considered to be largely “junk” DNA, lacking function. However, the fast-growing collection of genome-wide datasets describing chromatin features across increasing numbers of cell types has dramatically changed this view. These studies have started to reveal the organizational complexity of mammalian genomes, and it is at present speculated that ∼40% to 80% of the genome shows a biochemical signature that could imply functional relevance.1-3 Transcriptional enhancers represent a critical component of this noncoding regulatory genome as they bestow a unique identity on cells by establishing cell type-specific spatio-temporal gene expression patterns.4,5 In line with their essential roles in transcriptional regulation, numerous recent studies have causally linked aberrant enhancer function to human disorders and phenotypic variation, further demonstrating the important roles played by transcriptional enhancers in human biology.6-19 Through this review article, we aim to provide a concise update on new insights obtained in the last few years concerning the molecular mechanisms by which regulatory elements regulate gene expression, often over large genomic distances, and how disruption of these processes can contribute to the development of human disease. We will also discuss emerging therapeutic strategies aimed at manipulating the function of enhancers for the treatment of human genetic disorders.
Transcriptional regulation by enhancers is often a long-distance event
Many thousands of potential enhancers have been identified in the human genome,1 of which thousands are active in a given cell type.20-22 Enhancers are often localized at large distances from the genes they regulate, with an estimated median enhancer-target gene distance of 120 kb,23 although extreme cases of >1 Mb have been documented.8,12,24 They can be positioned both intragenic and intergenic, or even in nonrelated genes, and do not necessarily regulate transcription of the nearest gene.25 Enhancers regulate genes over large genomic distances via chromatin looping, bringing distal enhancers and the regulatory protein complexes that bind them in close nuclear proximity to their target genes. Chromatin loop formation has therefore been shown to be a better predictor of enhancer target genes than enhancer-gene linear proximity25 (although it is important to note that chromatin looping does not functionally connect enhancers and promoters per se).
Well-studied examples of such long-range gene regulation in the hematopoietic system are the erythroid globin,26-28 BCL11A,13 Myb,29 and Kit30 gene loci. Gene regulatory chromatin-looping events are thought to be dynamic and actively modulated during differentiation to accommodate for the changes in target gene expression necessary during development and cellular differentiation, although a recent study in Drosophila indicates that enhancer-promoter loops may be remarkably stable during development.31
Because of the high degree of complexity and specificity required for enhancer gene communication, chromosome conformation needs to be highly organized. A substantial body of evidence suggests that the determinants of promoter enhancer specificity can be very diverse,32-34 ranging from transcription factors (TFs),30,35-38 chromatin modifying proteins,39 or so-called “architectural” proteins (ie, CTCF, Cohesin, and Mediator)40-44 to noncoding RNAs (eg, enhancer RNAs and long noncoding RNAs).45,46 Enhancer-promoter interactions are promoted by the confinement of such interactions to chromosome structural domains called topological-associated domains (TADs), which partition chromosomes into discrete submegabase- to megabase-sized domains.47-49 This observation suggests a “loops within loops” model, where TADs provide a structural environment that prevents enhancer promiscuity.50 Other studies suggest that active genes regulated by similar TF complexes tend to cluster in the nuclear space or even colocalize in specific nuclear foci referred to as transcription factories.50-54 Although the occurrence of (active) gene movement toward relatively static transcription factories is presently still under debate,54,55 it is clear that different scales of genome folding are intimately linked to transcription regulation by allowing proper enhancer-gene contacts, placing enhancers and genome spatial organization at the heart of transcription.
Spatial genome organization and human disease
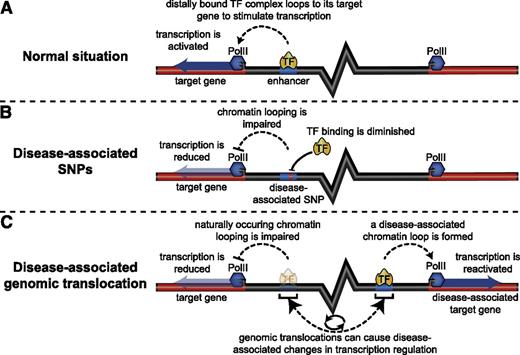
The majority of identified disease-associated genomic mutations and polymorphisms are located in noncoding DNA regions, often colocalizing with potential regulatory sequences.1 For several examples, single nucleotide polymorphisms have been shown to significantly influence long-range chromatin folding (Figure 1).14,15 In addition, cancer cells typically show massive structural and spatial chromosomal rearrangements that potentially displace regulatory elements from their native into an ectopic environment.6-11 As a consequence, novel long-range interactions can be established between normally unrelated enhancer-promoter pairs, leading to improper gene regulation and cellular transformation (Figure 1).7,56-58 An elegant study by the Delwel laboratory recently reported a prime example of how aberrant enhancer “rewiring” can cause disease.7 They investigated acute myeloid leukemia (AML) cells bearing inv(3)/t(3;3) chromosomal rearrangements, which are characterized by the transpositioning of a GATA2 enhancer into the EVI1 stem cell regulator locus. This chromosomal abnormality causes leukemia through the inappropriate long-range activation of EVI1 expression by the ectopic GATA2 enhancer, possibly reinforced by the accompanying reduction of GATA2 expression. Studies by the Yamamoto group using a transgenic mouse model of inv(3) AML further confirmed these observations.58 Another recent study investigated the molecular basis of pediatric medulloblastoma, in which complex chromosomal rearrangements activate the GFI1 and GFI1B oncogenes by placing them under the transcriptional control of unrelated enhancer elements,56 an event the authors referred to as “enhancer hijacking.” These studies highlight the potential pathological impact of regulatory element displacement in human disease, underscoring the value of investigating spatial genomic organization when dissecting the molecular events associated with cancer.
Genomic alterations can affect gene regulation via chromosome conformation. (A) Chromatin folding plays an essential role in transcriptional control by distally located regulatory elements. (B) Disease-associated single nucleotide polymorphisms located in distal regulatory elements can influence long-range chromatin interactions through their detrimental effect on the recruitment of TF complexes (eg, by destroying a TF binding motif), resulting in reduced expression of the target gene. (C) Chromosomal aberrations (eg, translocations) can relocate distal enhancers near a disease-associated gene, leading to the formation of pathological long-range chromatin interactions that ectopically activate the expression of this gene.
Genomic alterations can affect gene regulation via chromosome conformation. (A) Chromatin folding plays an essential role in transcriptional control by distally located regulatory elements. (B) Disease-associated single nucleotide polymorphisms located in distal regulatory elements can influence long-range chromatin interactions through their detrimental effect on the recruitment of TF complexes (eg, by destroying a TF binding motif), resulting in reduced expression of the target gene. (C) Chromosomal aberrations (eg, translocations) can relocate distal enhancers near a disease-associated gene, leading to the formation of pathological long-range chromatin interactions that ectopically activate the expression of this gene.
Therapeutical targeting of enhancers
Because of their key role in human disease, targeting these newly established disease-associated long-range interactions has become an attractive therapeutic strategy. Recent advances in genome editing technologies, such as using Zinc-finger nucleases (ZnFs), transcription activator-like effector nucleases (TALENs), or the CRISPR/Cas9 system (see Gaj et al.59 and Gupta and Musunuru60 for comprehensive reviews), make targeted enhancer-modifying strategies feasible and open up exciting new avenues for the therapeutic manipulation of genome topology.
In the abovementioned study by Gröschel and colleagues,7 excision of the oncogenic GATA2 enhancer from the AML genome, using either TALEN- or CRISPR/Cas9-mediated genome editing, induced in vitro growth arrest and differentiation of the edited leukemic cells, demonstrating the promising potential of hijacked oncogenic enhancers as therapeutic targets.
Enhancer activity is often highly cell type specific, and even widely expressed genes seem to possess tissue-specific enhancers driving their expression.61 This cell type-specific nature of enhancers makes them suitable targets for tissue-specific modulation of gene expression. Targeting enhancers (rather than promoters or the gene products themselves) offers the advantage of allowing cell type-specific silencing of gene expression (Figure 2). Such a strategy would even allow reducing the levels of currently undruggable proteins, including TFs.62 This principle was first explored by Bauer et al, using genome-editing technology to delete an erythroid-specific enhancer of the BCL11A TF gene.13 BCL11A is widely recognized as an important therapeutic target for the treatment of β-thalassemias and sickle cell anemia (the β-hemoglobinopathies), 2 common erythroid genetic disorders caused by, respectively, a quantitative or qualitative defect in adult hemoglobin production.63,64 In erythroid cells, BCL11A plays an important role in β-globin gene regulation, and its depletion in adult erythroid cells leads to a strong reactivation of fetal (β-like) γ-globin gene expression, which can efficiently compensate for the low abundant or defective adult hemoglobin.65-67 However, as BCL11A is a widely expressed TF and has been implicated in lymphomagenesis,68,69 targeting the protein itself remains problematic. In erythroid cells, BCL11A expression is controlled by intronic enhancers located 55 to 62 kb downstream of the transcription start site.13 Targeted deletion of these enhancers dramatically reduces BCL11A expression specifically in erythroid cells, showing that targeting enhancers allows for the tissue-specific silencing of broadly expressed genes. Other genome editing-based strategies to manipulate enhancer biology can be envisioned, such as the specific targeting of repressor (domain) fusion proteins to disease-associated enhancers. Mendenhall and colleagues pioneered this approach by fusing transcription activator–like (TAL) effector repeat domains to the LSD1 histone demethylase. Targeting this fusion protein to the genome efficiently reduced enhancer activity, resulting in the downregulation of nearby genes.70
Enhancer targeting strategies with proven therapeutical potential. (A) Deletion of enhancers can cell type specifically modulate disease-associated gene expression. (B) Artificial tethering of TFs to regulatory elements can restore disease-impaired chromatin looping, leading to reactivation of gene expression. Similar tethering strategies can also be used to artificially reactivate naturally silenced genes for therapeutic use74 or for enhancer silencing by tethering inhibitory proteins to the enhancer.70
Enhancer targeting strategies with proven therapeutical potential. (A) Deletion of enhancers can cell type specifically modulate disease-associated gene expression. (B) Artificial tethering of TFs to regulatory elements can restore disease-impaired chromatin looping, leading to reactivation of gene expression. Similar tethering strategies can also be used to artificially reactivate naturally silenced genes for therapeutic use74 or for enhancer silencing by tethering inhibitory proteins to the enhancer.70
Manipulating the loop: artificial enhancer-hijacking for therapeutic purposes?
As stated before, TFs are involved in establishing and stabilizing long-range chromatin interactions. The non-DNA binding adaptor protein LDB1 is required for the development of multiple tissues, including the hematopoietic system.71,72 LDB1 assembles a multiprotein TFC complex in erythroid cells, containing 2 essential DNA-binding TFs GATA1 and TAL1, and LDB1 is required for enhancer-promoter looping at the β-globin and Myb loci.29,36,37,73 A direct demonstration that LDB1 is the critical complex component mediating chromatin looping came from an elegant study by the Blobel laboratory.73 During erythropoiesis, the LDB1 TF complex binds the β-globin gene promoter and upstream locus control region (LCR) to achieve LCR promoter looping and high-level globin gene expression.36,37 In GATA1-deficient erythroid progenitors, LDB1 is only targeted to the LCR (via its interaction with TAL1) but not to the β-globin promoter. In these cells, long-range LCR-promoter interactions are absent, and the β-globin genes are therefore not expressed. Artificial ZnF-mediated tethering of LDB1, or the LDB1 dimerization domain only, to the β-globin promoter in these GATA1-deficient cells was shown to be sufficient for establishing the LCR-promoter loop, resulting in a (partial) activation of β-globin gene expression.73
This “forced-looping” strategy was then used in an attempt to reactivate fetal γ-globin gene expression in adult red blood cells as a potential new strategy for the treatment of β-hemoglobinopathies.74 Deng et al showed that forced chromatin looping of the β-globin LCR to the developmentally silenced fetal γ-globin gene reactivates its expression in cultured adult erythroid cells. This time, they manipulated local long-range chromatin interactions by targeting the ZnF-LDB1 dimerization domain fusion-protein to the human γ-globin gene promoter. Importantly, γ-globin expression reached levels that would be sufficient to significantly ameliorate the clinical disease course of β-hemoglobinopathy patients.75 This study provides the first proof of principle that long-range chromatin interactions can be artificially controlled—potentially even for therapeutic purposes—providing an invaluable addition to the ever-growing genomics toolbox at our disposal (Figure 2).
Clinical enhancer targeting: remaining challenges
Despite the attractiveness of enhancer targeting as a potential broadly applicable therapeutic approach, several important challenges need to be faced before such strategies become clinically applicable. Whereas ZnF-LDB1-based reactivation of γ-globin did not appear to have a major impact on the majority of a selected set of erythroid genes controlled by the endogenous LDB1 complex, the influence of overexpressed engineered fusion proteins on the activity of endogenous regulatory protein complexes and the genes they regulate needs to be rigorously tested. Similarly, the off-target effects of genomic engineering technologies are still under intense investigation, and several studies have highlighted the need for more optimized targeting strategies.76,77 In addition, because of the important role of regulatory elements in organizing spatial genome topology, potential side effects of (even small) genomic alterations on local three-dimensional chromosomal organization and gene regulation will have to be thoroughly investigated. Finally, there is the considerable challenge of efficient and specific delivery of the genome editing constructs to the target cell. With a few exceptions,78,79 this is still only feasible using ex vivo cultured (stem) cells that can be transplanted back into the recipient.80 Major technological developments in this area are required if the current genome editing methods are to be used for systemic therapy in vivo.
Conclusion
Enhancers that regulate gene expression over large distances by chromatin looping processes are critical for proper development and tissue homeostasis. Recent progress has clearly shown that the genetic disruption of enhancer function plays a widespread and important role in human phenotypic variation, disease susceptibility, and even disease etiology. As new technologies that allow the targeted manipulation of regulatory elements develop at an astonishing rate, we expect therapeutic strategies aimed at intervening with disease-associated enhancer-gene communication or at establishing therapeutically beneficial enhancer-gene interactions to become feasible in the future.
Acknowledgments
The authors thank the members of the Grosveld and Soler laboratories for helpful discussions. The authors apologize to the colleagues whose work could not be cited due to space limitations.
Authorship
Contribution: All authors contributed to the writing of the paper and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R.S. is Centre for Genomic Regulation, Barcelona, Spain.
Correspondence: E. Soler, INSERM UMR967, CEA/DSV/iRCM, Laboratory of Molecular Hematopoiesis, 92265, Fontenay-aux-Roses, France; e-mail: eric.soler@cea.fr.