Abstract
The past 40 years have witnessed major advances in defining the cytogenetic aberrations, mutational landscape, epigenetic profiles, and expression changes underlying hematological malignancies. Although it has become apparent that acute myeloid leukemia (AML) is highly heterogeneous at the molecular level, the standard framework for risk stratification guiding transplant practice in this disease remains largely based on pretreatment assessment of cytogenetics and a limited panel of molecular genetic markers, coupled with morphological assessment of bone marrow (BM) blast percentage after induction. However, application of more objective methodology such as multiparameter flow cytometry (MFC) has highlighted the limitations of morphology for reliable determination of remission status. Moreover, there is a growing body of evidence that detection of subclinical levels of leukemia (ie, minimal residual disease, MRD) using MFC or molecular-based approaches provides powerful independent prognostic information. Consequently, there is increasing interest in the use of MRD detection to provide early end points in clinical trials and to inform patient management. However, implementation of MRD assessment into clinical practice remains a major challenge, hampered by differences in the assays and preferred analytical methods employed between routine laboratories. Although this should be addressed through adoption of standardized assays with external quality control, it is clear that the molecular heterogeneity of AML coupled with increasing understanding of its clonal architecture dictates that a “one size fits all” approach to MRD detection in this disease is not feasible. However, with the range of platforms now available, there is considerable scope to realistically track treatment response in every patient.
Rationale for detection of minimal residual disease in acute myeloid leukemia
The pioneering work of investigators such as Janet Rowley established that acute myeloid leukemia (AML) is a genetic disease. Even at the resolution of conventional cytogenetics, it is clear that AML is highly heterogeneous at the molecular level, with more than 100 recurring balanced chromosomal rearrangements identified to date, encompassing biologically and prognostically distinct disease entities (reviewed in Grimwade and Mrózek1 ). Although cytogenetics continues to provide the framework for risk stratification used to guide the management of AML, there has been inconsistency in the classification systems employed by different trial groups, with variations having a potential bearing on transplant decisions in individual patients.1 In a large meta-analysis by Cornelissen and colleagues2 considering the relationship between cytogenetic risk groups and outcome, a predicted relapse rate exceeding 35% was proposed as the most informative disease-related cutoff to guide transplant decisions in first remission.
After advances in sequencing technology that led to the discovery of a number of novel recurrent mutational targets in AML, there has been increasing interest in the use of molecular profiling using targeted sequencing panels to further refine risk stratification.3,4 Nevertheless, beyond those abnormalities distinguishable by cytogenetics, to date only a very limited number of molecular markers (ie, mutations involving NPM1, CEBPA, FLT3-ITD, cKIT) have gained acceptance as being of clinical relevance, as recognized in widely used disease guidelines.5,6 However, the seminal Cancer Genome Atlas consortium study that involved sequencing 200 AML genomes covering the cytogenetic spectrum of the disease has provided evidence that the average AML case harbors more than 10 mutations, with in excess of 200 genes being recurrent mutation targets and more than 1 000 genes found to be mutated in at least 1 of the 200 cases analyzed.7 This degree of complexity presents a major challenge for defining individual genetic abnormalities or combinations of markers that provide significant independent prognostic information and establish their respective relationships to other pretreatment characteristics known to influence outcome (eg, patient age, presenting white blood cell count [WBC], secondary AML).
Although pretreatment characterization of AML based on cytogenetic analysis and molecular profiling can evidently distinguish broad subgroups of patients with relatively favorable, unfavorable, or intermediate prognosis, major drawbacks are that it lacks the capacity to pinpoint precisely which patients can be cured with conventional chemotherapy alone and may therefore be spared the toxicity associated with an unnecessary transplant, and, conversely, to reliably identify those most likely to benefit from early transplantation. Importantly, although cytogenetics and molecular profiling can provide a snapshot of the genetic makeup of each case of leukemia, they are collectively insufficient to fully capture other parameters that may have a significant bearing on disease outcome. These may in some instances reflect differences in the cellular origins of the leukemia and sensitivity of the leukemic clone to therapy, which may be influenced by the biology of the neoplastic cells, interaction with the marrow microenvironment, and interpatient variation in drug availability and metabolism, which may also be genetically determined.
Despite the major advances in understanding the molecular pathogenesis of AML, assessment of bone marrow (BM) morphology remains the standard approach to gauge treatment response in routine care and in the clinical trial setting, and is used to inform decisions concerning allogeneic transplantation. However, determination of blast percentage by light microscopy is hampered by limited sensitivity and interobserver variability. Indeed, a recent study conducted in a large cohort of pediatric AML patients (n = 203) reported marked discrepancies in evaluation of remission status as determined by standard morphology and flow cytometry.8 Some patients considered to be in morphological remission had in excess of 5% leukemic blasts by flow cytometry. Conversely, 67% of patients classified on morphological grounds to have a partial response (5% to 15% blasts) and 26% of those classified as having resistant disease (>15% blasts) actually had an excellent response according to flow cytometry, with no detectable minimal residual disease (MRD).8 Other studies have shown similar discrepancies between morphological and flow cytometric assessment, with results from the latter having the greatest impact on prognosis.9,10 The obvious caveat to this is the effect of sample hemodilution on the accuracy of flow cytometric results.
Taking into account the limitations of pretreatment characterization of the leukemic clone to reliably predict outcome in individual patients relating in part to the molecular complexity highlighted previously, together with the problems in using morphology to reliably assess remission status, there is clearly a very strong rationale for use of MRD detection methods to provide a more objective assessment of treatment response to develop a more individualized approach to management. This review will consider the various platforms available at present or in the near future, take account of their relative advantages and limitations, and discuss the most informative approach depending on the subtype of AML and clinical scenario, recognizing the challenges for the laboratory involved in the successful delivery of MRD-directed therapy.
Flow cytometry
The accumulated evidence from studies of multiparameter flow cytometric MRD (MFC-MRD) assessment in AML (reviewed in Buccisano et al11 and Kern et al 12 ), including the most recent,9,10,13 leaves little doubt that this method of MRD detection can be used to risk-stratify both younger and older patients at treatment time points, including predicting outcome postallogeneic stem cell transplantation (reviewed in Buckley et al14 and Campana and Leung15 ). The prognostic impact of MFC-MRD is strong enough to have emerged despite study differences in the MFC assays and the limitations of now-outdated restricted antibody panels. Its clinical value as a biomarker to inform therapy is therefore difficult to ignore, particularly with its applicability to the majority of patients. However, it is also apparent that these results have all been generated by and therefore are dependent on a centralized laboratory approach with a track record of analysis expertise. Because replication of this requires the organization of specialized core laboratories, the implementation of flow cytometric measurement of MRD has lagged behind that of real-time quantitative PCR.
Assay approaches
Continued improvements in multiparameter flow technology and reagents enable simultaneous measurement of up to 20 antigens/markers per cell compared with 3 in initial AML MFC-MRD reports,16 allowing higher resolution detection per tube of BM cells. This capability is potentially further increased by mass cytometry (up to 45 parameters, but with slower cell acquisition). Visualization and analysis of immunophenotypic data may be transformed in the near future by clinical use of automated analysis algorithms such as SPADE17 and ViSNE18 but at present, defining and detecting abnormal leukemic cells from the data produced by flow cytometry still rely on manual inspection of cells by 2 parameters at a time on biaxial plots with multiple gating steps of selected cell populations. This MFC-MRD analysis becomes more complex with increased numbers of simultaneous fluorochrome parameters (ie, colors), as do the technical issues (eg, compensation, fluorescence overlap reducing signal sensitivity); hence, most current AML MFC-MRD antibody panels use 6 to 10 colors.
There are 2 main analysis strategies for detecting MFC-MRD. The first, using a screening antibody panel (Figure 1), selects antigen combinations at diagnosis (termed leukemia-associated immunophenotypes [LAIPs]), each displayed by at least 5% to 10% of leukemic cells (initially gated by blast markers) and sufficiently aberrant to be present on less than 0.1% to 0.01% of normal nucleated BM cells, so that in an adequate sample, an MRD detection sensitivity threshold of 10−3 to 10−4 (1 in 1 000 to 1 in 10 000 cells) can be achieved. The most sensitive/specific LAIPs (reducing MRD false positives) will identify leukemic cells in “empty spaces”—regions defined on dot plots (after applying other parameter-based gates) that contain no or very few cells in control BM samples (including “stressed” regenerating normal marrows). Subsequent MFC-MRD monitoring tracks these diagnostic LAIPs (sometimes using tailored antibody combinations), with a cluster of 20 cell events in the LAIP gate being potentially sufficient to identify MRD—resulting in a maximum sensitivity of between 10−4 and 10−5 when 500 000 nucleated cells are examined. Identifying specific LAIPs at diagnosis may be improved by incorporation of more colors, thereby allowing the addition of further simultaneous markers required to define an abnormal cell. However, this is a double-edged sword because instability of even 1 of the LAIP markers after treatment increases the risk of an MRD false negative. Experience of which phenotypic changes are more common (such as a shift to a more immature phenotype)19 allows selection of LAIPs that are the most robust to monitor.
Outline of antibody groups used in panels for identification of AML-aberrant immunophenotypes (both for LAIP and “different-from-normal” approaches) and subsequent residual disease monitoring. Core markers are those selected for the backbone of the panel to identify myeloid blast populations. These are combined with markers from lymphoid/myelomonocytic maturation groups (megakaryocytic markers and NG2 [for MLL-rearranged AML] are not included but are more useful in pediatric AML). A stem cell combination may be included (as in the United Kingdom [UK] National Cancer Research Institute [NCRI] AML trial panel) to detect potential immunophenotypic LSC. Most sensitive, robust, aberrant phenotypes include cross-lineage expression and CD34+ human leukocyte antigen (HLA) DR weak/negative. Some aberrant phenotypes may be sensitive (by testing detection by serial dilution in normal marrow) but less stable/useful in follow-up samples. Blue, antibody marker; bold blue, marker in NCRI AML trial panel; red, type of aberrant phenotype potentially detected by marker group.
Outline of antibody groups used in panels for identification of AML-aberrant immunophenotypes (both for LAIP and “different-from-normal” approaches) and subsequent residual disease monitoring. Core markers are those selected for the backbone of the panel to identify myeloid blast populations. These are combined with markers from lymphoid/myelomonocytic maturation groups (megakaryocytic markers and NG2 [for MLL-rearranged AML] are not included but are more useful in pediatric AML). A stem cell combination may be included (as in the United Kingdom [UK] National Cancer Research Institute [NCRI] AML trial panel) to detect potential immunophenotypic LSC. Most sensitive, robust, aberrant phenotypes include cross-lineage expression and CD34+ human leukocyte antigen (HLA) DR weak/negative. Some aberrant phenotypes may be sensitive (by testing detection by serial dilution in normal marrow) but less stable/useful in follow-up samples. Blue, antibody marker; bold blue, marker in NCRI AML trial panel; red, type of aberrant phenotype potentially detected by marker group.
The second analysis approach, initially validated by the Children’s Oncology Group,9,20 circumvents the problems of false negatives from phenotypic shifts and emerging subclones by using a fixed antibody panel (also based on combinations in Figure 1) and an analysis that screens for established immunophenotypic profiles to distinguish abnormal leukemic cells (including more mature subpopulations) from normal cells irrespective of diagnostic leukemic immunophenotype. This is termed “different-from-normal” MFC-MRD, perhaps confusingly, because diagnostic LAIPs in the first approach are also different from normal. This type of MFC-MRD assay has been successfully applied to allogeneic transplant patients,21 in which there frequently are insufficient data from referring hospitals to monitor diagnostic LAIPs and there may be no known leukemia-specific genetic marker (Figure 2). One potential consideration of this approach is that different-from-normal phenotypes, particularly in more mature myeloid cells, may result from preleukemic clones22-24 and these, although chemoresistant, will have a lower early relapse risk compared with leukemic clones. The implications of this for relapse prediction is likely to be particularly relevant to older patients with their higher prevalence of background myelodysplasia (MDS)/preleukemic clones and also apply to the LAIP assay since aberrant blast immunophenotypes have been observed in low risk MDS.25
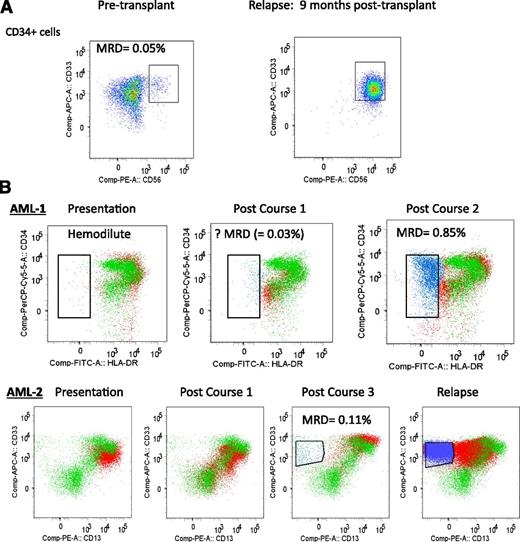
Examples of detecting MRD with “different-from-normal approach” applied to myeloblast population. (A) AML patient undergoing allogeneic stem cell transplant with no prior flow cytometric data available to identify diagnostic LAIP; however, pretransplant bone marrow CD34+ myeloblasts (defined by gating using CD34+/CD117+/CD45/SSC/FSC parameters) include a clearly aberrant CD34+CD33+CD56+ leukemic population and therefore are MRD-positive (0.05% of BM-nucleated cells). This patient relapsed with the same aberrant phenotype. (B) Examples of 2 AML patients (AML-1, AML-2) monitored for MRD during chemotherapy using data overlay with a control sample to detect “different-from-normal” blast subpopulations in CD117+ myeloblasts (defined by gating using CD117+/CD45/ SSC/FSC parameters). Green, CD117+ myeloblasts of control bone marrow; red, CD117+ myeloblasts of patient’s bone marrow; blue, emerging aberrant leukemic subpopulation within empty space; empty space, region in which there are very few or no normal cells. (AML-1) CD34 vs HLADR plots shown of CD117+ blasts. Presentation sample was hemodilute with no definite LAIP (other markers not shown). Postcourse 1 sample had a small number of cells in an empty space (CD34+HLADRlow, in blue) but categorized as insufficient to define as MRD without a diagnostic LAIP; however, the postcourse 2 sample had obvious MRD within the same empty space. (AML-2) CD33 vs CD13 plot shown of CD117+ blasts. Change in leukemic immunophenotype with MRD postcourse 3 from an emerging new aberrant subpopulation in an empty space (CD33+CD13low, in blue). This patient relapsed with the same aberrant phenotype but the diagnostic LAIPs were not present (including from other markers not shown).
Examples of detecting MRD with “different-from-normal approach” applied to myeloblast population. (A) AML patient undergoing allogeneic stem cell transplant with no prior flow cytometric data available to identify diagnostic LAIP; however, pretransplant bone marrow CD34+ myeloblasts (defined by gating using CD34+/CD117+/CD45/SSC/FSC parameters) include a clearly aberrant CD34+CD33+CD56+ leukemic population and therefore are MRD-positive (0.05% of BM-nucleated cells). This patient relapsed with the same aberrant phenotype. (B) Examples of 2 AML patients (AML-1, AML-2) monitored for MRD during chemotherapy using data overlay with a control sample to detect “different-from-normal” blast subpopulations in CD117+ myeloblasts (defined by gating using CD117+/CD45/ SSC/FSC parameters). Green, CD117+ myeloblasts of control bone marrow; red, CD117+ myeloblasts of patient’s bone marrow; blue, emerging aberrant leukemic subpopulation within empty space; empty space, region in which there are very few or no normal cells. (AML-1) CD34 vs HLADR plots shown of CD117+ blasts. Presentation sample was hemodilute with no definite LAIP (other markers not shown). Postcourse 1 sample had a small number of cells in an empty space (CD34+HLADRlow, in blue) but categorized as insufficient to define as MRD without a diagnostic LAIP; however, the postcourse 2 sample had obvious MRD within the same empty space. (AML-2) CD33 vs CD13 plot shown of CD117+ blasts. Change in leukemic immunophenotype with MRD postcourse 3 from an emerging new aberrant subpopulation in an empty space (CD33+CD13low, in blue). This patient relapsed with the same aberrant phenotype but the diagnostic LAIPs were not present (including from other markers not shown).
MFC-MRD assay sensitivity
MFC-MRD status postinduction not surprisingly appears most predictive for early relapses10,13 as well as a surrogate for overall survival. An experienced laboratory and an adequate sampling26 optimize assay sensitivity, but from published studies about one-third of younger patients without detectable MFC-MRD at predictive time points will relapse; these are the false negatives of the assay. These may result from the phenotypic changes discussed previously or emergence of initially minor subpopulations (less than 5% to 10% of blasts at presentation) excluded by the LAIP assay or may reflect chemoresistant leukemic cells that lack sufficiently specific immunophenotypic aberrancies, even if these are identified in other leukemic subpopulations. Although extensive antibody panel screens increase the chances of detecting rarer aberrancies, cost-effectiveness and sample size are practical considerations limiting their use. Alternatively the chemoresistant reservoir of MRD may predominate in extramedullary sites or consist of rare leukemic cells/leukemic stem cells (LSC) below the detection threshold of 10−4. Assays detecting LSC rather than bulk AML blast subpopulations may increase sensitivity and predictive value of MRD monitoring. Because functional xenograft assays for LSC are not clinically applicable, the alternative strategy is to monitor the frequency of candidate immunophenotypic populations enriched for LSC. Published assays have mainly focused on the CD34+CD38− compartment, which in normal BM contains hematopoietic stem cells, either using aberrant differential expression of non-hematopoietic stem cell markers27 similar to the LAIP/different-from-normal approach or measuring abnormal expansion of a CD34+CD38− stem/progenitor compartment previously functionally characterized to contain AML LSC.28 However, LSC are not restricted to this immunophenotypic subset in all patients, because AML blasts with LSC activity defined functionally by xenotransplant models have heterogenous surface marker profiles including those of CD34+CD38+ and sometimes CD34− populations.29-31 Flow cytometric assays to detect LSC in the CD34+CD38+ or CD34− compartments are in development.32 Recent data suggest that xenograft LSC frequency of different AML genetic subclones from presentation samples may not necessarily predict their emergence at relapse,23 underlining the need to evaluate the clinical relevance of any potential MFC LSC residual disease assay.
Standardization of MFC-MRD
Despite the powerful prognostic value of MFC-MRD, standardization and therefore comparability of results between laboratories remains problematic. Although MFC-MRD can be applied to most AML patients, this involves multiple aberrant antigen combinations detected by evolving antibody panels developed separately by laboratories. This is analogous to standardizing quantitative polymerase chain reaction (PCR) of multiple molecular targets with laboratories using different primers for each target. There is variation in the quantitation of MFC-MRD (ie, % of total nucleated cells vs % of CD45+ cells vs % of mononuclear cells) in published studies affecting cutoffs/thresholds for the lower limit of the quantitative range and the significance of MRD positivity at different treatment time points. There are also different approaches to defining MRD positivity/negativity, either by using cutoff values derived from combined data of multiple phenotypic aberrancies at postinduction/consolidation time points or by defining MRD positivity as any level of MRD detectable above the relevant aberrant combination sensitivity threshold.10,33 A recent study13 using the first approach tested different MFC-MRD cutoff values by relapse probability and showed significant differences in relapse for MFC-MRD detected above ∼0.05% of CD45+ cells post–induction cycles. The cutoff value was similar whether or not the MFC-MRD value incorporated a correction for LAIP frequency in the presentation sample and was not improved by using log reduction values. Although 0.1% of CD45+ cells is the most frequently applied lower limit of the quantitative range for combined LAIP data, this will inevitably increase the frequency of false negatives that might be detected by applying lower cutoffs for the more sensitive LAIPs as in the second approach. Integration of flow cytometric and genetic data will allow optimization of predictive cutoffs for specific LAIPs when these are associated with different genetic abnormalities—for example, aberrant CD7 in AML with CEBPA or FLT3-ITD mutations. Incorporation of a correction factor for hemodilution34 is another consideration for standardized reporting as well as reduction of false negatives.
It is likely that stronger standardization of MFC-MRD assays will evolve from networked centralized laboratories such as in European initiatives (eg, Feller et al35 ), taking into account the differences between pediatric and adult AML for the specificity and frequency of aberrancies. Improved selection and analysis of leukemic aberrancies could be achieved by Web-based access to international interlaboratory shared resources of the most useful robust LAIPs/different-from-normal profiles/analysis strategy, together with agreement to test newer combinations.36 Data from samples, together with control BM processed by standardized flow cytometric protocols in different laboratories, could be analyzed remotely, thereby accessing a core facility of appropriate expertise. It is feasible that further development of automated analysis algorithms combined with high-dimensional cytometry applied as a different-from-normal assay will provide both standardization and high resolution.
Real-time quantitative PCR
The development of these assays in the 1990s provided a major step forward in establishing standardized approaches for MRD detection in a range of leukemias. In AML, they can be applied in cases with chimeric fusion genes generated by balanced chromosomal rearrangements—for example, PML-RARA/t(15;17), RUNX1-RUNX1T1/t(8;21), CBFB-MYH11/(inv(16)/t(16;16), DEK-CAN(NUP214)/t(6;9), t(11q23)/MLL fusions, t(5;11)/NUP98-NSD1 or NPM1 mutations, collectively covering ∼60% of AML presenting in children and younger adults (Figure 3).1,37,38 For these assays, an RNA-based approach is used, first undertaking reverse transcription (RT) to generate complementary DNA before the quantitative PCR (qPCR) step. This allows a relatively limited panel of optimized standardized assays to be used, circumventing the need to characterize translocation breakpoints at the genomic level, which can be challenging and not realistic in routine laboratories.
Proportion of AML patients informative for MRD detection by RT-qPCR for leukemia-specific MRD targets (ie, fusion genes, NPM1 mutation) according to age.
Proportion of AML patients informative for MRD detection by RT-qPCR for leukemia-specific MRD targets (ie, fusion genes, NPM1 mutation) according to age.
A landmark in the standardization of this methodology for clinical implementation was the Europe Against Cancer (EAC) program, which established a framework for assessment and selection of optimal RT-qPCR assays through systematic parallel evaluation in an international network of expert laboratories.39 This involved design of a common primer and TaqMan probe for each respective MRD target (ie, fusion gene) to be used in conjunction with different variable primers to cover the common isoform types encountered in primary patient samples. Specific probes (rather than SYBR green I) were used to detect target amplicons to enhance assay sensitivity and specificity. Apart from establishing common protocols that included reaction conditions for all steps of the RT-qPCR procedure, a key achievement of the EAC program was the rigorous evaluation of a large panel of potential housekeeping genes to identify candidates with stable expression in normal peripheral blood (PB) and BM, similarly expressed across a range of leukemias and with comparable stability to that of the leukemic transcripts. This process identified ABL as the most reliable control gene.40 Although some laboratories prefer to use alternative housekeeping genes, there is no evidence that these are superior; therefore, consistent use of ABL for assay normalization would greatly facilitate comparison of MRD data between laboratories. Importantly, the EAC program also recommended that assays be run in triplicate wells, provided guidance concerning acceptable sample quality indicated by the level of housekeeping gene expression and established clear criteria used to define PCR positivity (ie, specific amplification in the MRD assay in at least 2 of 3 replicate wells with average cycle threshold value ≤40).
Although some laboratories still use conventional RT-PCR with nested primers for MRD detection, this approach has a number of limitations and should be abandoned in favor of RT-qPCR, which is more reliable and readily standardized. Performance of the same RT-qPCR platform is highly reproducible between laboratories, turnaround time is more rapid, and risk of PCR contamination is substantially reduced. A further key advantage of RT-qPCR is the capacity to quantify an independent housekeeping gene in parallel, enabling suboptimal follow-up samples that could potentially have given rise to false-negative PCR results to be identified and that cannot be reliably distinguished by standard nested RT-PCR assays. Importantly, qualitative end-point assays lack the capacity to measure the absolute level of leukemic transcripts or determine whether they are rising or falling, which is invaluable information for clinical decision-making. The EAC program laid the groundwork for defining optimal MRD monitoring schedules, which need to take into account the maximal assay sensitivity (indicated by level of expression of leukemic transcripts relative to the control gene in the blast population defined at diagnosis), the most appropriate sample source (PB vs BM) and the typical kinetics of disease relapse (Figure 4).
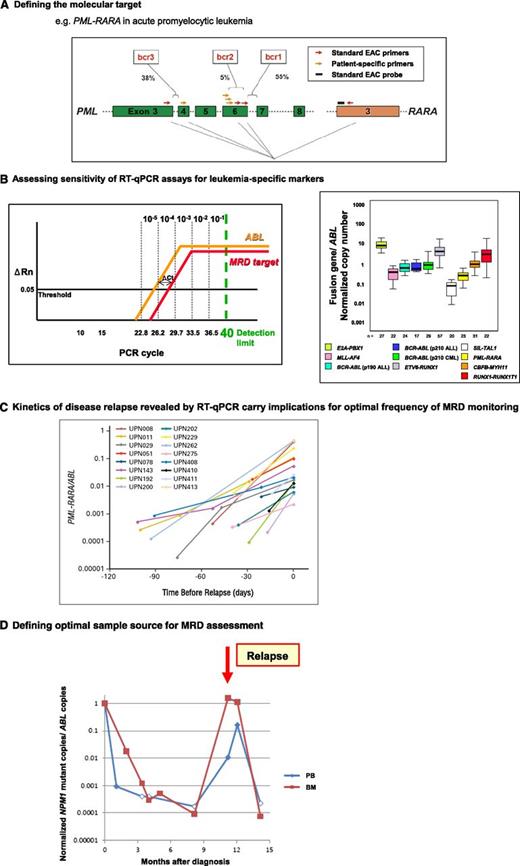
Development of leukemia-specific RT-qPCR assays to track treatment response is dependent upon molecular characterization of diagnostic material to determine the most appropriate assay, with MRD monitoring strategies informed by maximal achievable sensitivity, optimal sample type, and typical kinetics of disease relapse. (A) Analysis of diagnostic material is critical to determine the appropriate assay primer and probe set to detect MRD in any given patient because of heterogeneity in chromosomal breakpoints (eg, PML-RARA, CBFB-MYH11, MLL fusions) or mutation type (NPM1). For example, in ∼5% of acute promyelocytic leukemia cases, the standard EAC assays are not suitable because of occurrence of rarer breakpoints within the PML locus requiring design of patient-specific forward primers to be used in conjunction with the standard EAC probe and reverse primer (both located in RARA).44 Figure panel adapted from Grimwade et al46 with permission. (B) A key determinant of the sensitivity for MRD detection is the relative level of expression of the leukemia-specific transcript (ie, fusion gene, NPM1 mutant) as indicated by comparison with that of an endogenous control gene (eg, ABL). This can be measured as the difference in the number of PCR cycles (ΔCt) to detect fluorescence above background from amplification of the leukemic transcript and the control gene at the threshold (set at 0.05 according to EAC criteria39 ); see left panel. The detection limit of PCR is taken as 40 cycles (equivalent to ∼1 copy), with 1-log being equivalent to 3.45 cycles, as determined from the slope of the plasmid standard curve. Assuming ABL amplification at cycle threshold (Ct) value of 24, the observed Ct value for amplification of the leukemic target in blasts at diagnosis indicates the maximal theoretical sensitivity for detection of MRD in that particular patient. The Ct value of the MRD target equating with a given level of sensitivity (10−1 to 10−5) is marked based on an ABL Ct value of 24. For example, MRD can be detected at a sensitivity of at least 1 in 104, where ΔCtTarget-ABL is ≤2.2. Detection of MRD at a sensitivity of 1 in 105 is possible when the MRD target is more highly expressed than ABL, with a ΔCt of −1.2. ΔRn, normalized reporter signal (change in fluorescence intensity). Figure panel adapted from Freeman et al26 with permission. Examination of diagnostic BM samples from primary leukemia samples using standardized assays developed within the EAC program demonstrates marked variation in the level of leukemic transcripts both between and within different molecular subsets, which impacts on the sensitivity to detect MRD in any given patient (right panel). Figure adapted from Gabert et al39 with permission. (C) Apart from maximal assay sensitivity, a further parameter to take into account in determining MRD sampling schedules is the kinetic of disease relapse. For example, in APL, the median increment in PML-RARA fusion transcripts is ∼1-log/month. Reproduced from Grimwade et al44 with permission. (D) Parallel tracking of MRD status by RT-qPCR in PB and BM in a patient with NPM1 mutant AML, with filled and unfilled data points indicating that disease transcripts were detectable or undetectable, respectively. For PCR-negative samples, data points are plotted according to the maximal sensitivity afforded by the follow-up sample based on the respective level of ABL control gene expression and taking into account the difference in expression between the NPM1 mutant allele and ABL in leukemic cells at diagnosis (ΔCtNPM1mut-ABL), as described in panel B. In this patient, rapid PCR negativity was achieved in the PB. However, serial BM samples afforded greater sensitivity, revealing that the patient failed to achieve molecular remission after frontline therapy, with relapse preceded by a rapid rise in NPM1 mutant transcripts. The PB MRD assay only converted to PCR positivity at the time of diagnosis of clinical relapse.
Development of leukemia-specific RT-qPCR assays to track treatment response is dependent upon molecular characterization of diagnostic material to determine the most appropriate assay, with MRD monitoring strategies informed by maximal achievable sensitivity, optimal sample type, and typical kinetics of disease relapse. (A) Analysis of diagnostic material is critical to determine the appropriate assay primer and probe set to detect MRD in any given patient because of heterogeneity in chromosomal breakpoints (eg, PML-RARA, CBFB-MYH11, MLL fusions) or mutation type (NPM1). For example, in ∼5% of acute promyelocytic leukemia cases, the standard EAC assays are not suitable because of occurrence of rarer breakpoints within the PML locus requiring design of patient-specific forward primers to be used in conjunction with the standard EAC probe and reverse primer (both located in RARA).44 Figure panel adapted from Grimwade et al46 with permission. (B) A key determinant of the sensitivity for MRD detection is the relative level of expression of the leukemia-specific transcript (ie, fusion gene, NPM1 mutant) as indicated by comparison with that of an endogenous control gene (eg, ABL). This can be measured as the difference in the number of PCR cycles (ΔCt) to detect fluorescence above background from amplification of the leukemic transcript and the control gene at the threshold (set at 0.05 according to EAC criteria39 ); see left panel. The detection limit of PCR is taken as 40 cycles (equivalent to ∼1 copy), with 1-log being equivalent to 3.45 cycles, as determined from the slope of the plasmid standard curve. Assuming ABL amplification at cycle threshold (Ct) value of 24, the observed Ct value for amplification of the leukemic target in blasts at diagnosis indicates the maximal theoretical sensitivity for detection of MRD in that particular patient. The Ct value of the MRD target equating with a given level of sensitivity (10−1 to 10−5) is marked based on an ABL Ct value of 24. For example, MRD can be detected at a sensitivity of at least 1 in 104, where ΔCtTarget-ABL is ≤2.2. Detection of MRD at a sensitivity of 1 in 105 is possible when the MRD target is more highly expressed than ABL, with a ΔCt of −1.2. ΔRn, normalized reporter signal (change in fluorescence intensity). Figure panel adapted from Freeman et al26 with permission. Examination of diagnostic BM samples from primary leukemia samples using standardized assays developed within the EAC program demonstrates marked variation in the level of leukemic transcripts both between and within different molecular subsets, which impacts on the sensitivity to detect MRD in any given patient (right panel). Figure adapted from Gabert et al39 with permission. (C) Apart from maximal assay sensitivity, a further parameter to take into account in determining MRD sampling schedules is the kinetic of disease relapse. For example, in APL, the median increment in PML-RARA fusion transcripts is ∼1-log/month. Reproduced from Grimwade et al44 with permission. (D) Parallel tracking of MRD status by RT-qPCR in PB and BM in a patient with NPM1 mutant AML, with filled and unfilled data points indicating that disease transcripts were detectable or undetectable, respectively. For PCR-negative samples, data points are plotted according to the maximal sensitivity afforded by the follow-up sample based on the respective level of ABL control gene expression and taking into account the difference in expression between the NPM1 mutant allele and ABL in leukemic cells at diagnosis (ΔCtNPM1mut-ABL), as described in panel B. In this patient, rapid PCR negativity was achieved in the PB. However, serial BM samples afforded greater sensitivity, revealing that the patient failed to achieve molecular remission after frontline therapy, with relapse preceded by a rapid rise in NPM1 mutant transcripts. The PB MRD assay only converted to PCR positivity at the time of diagnosis of clinical relapse.
Clinical evaluation of RT-qPCR assays
PML-RARA in APL.
Use of MRD monitoring to inform clinical management has been best established in acute promyelocytic leukemia (APL), in which achievement of molecular remission in BM (ie, PML-RARA transcripts undetected at a sensitivity of at least 1 in 104) is a prerequisite for cure, leading to introduction of MRD assessment as a component of the standard response criteria in this subtype of leukemia.41,42 Although conventional nested RT-PCR assays have been widely used for MRD assessment in APL, as described previously, these assays have their limitations and have been superseded by quantitative assays. The standardized EAC PML-RARA RT-qPCR assay has been shown to improve MRD detection rates compared with conventional nested RT-PCR and has been extensively validated within the context of clinical trials.42-45 In a large UK study involving 406 patients treated with all-trans retinoic acid + anthracycline-based therapy, mostly in the MRC AML15 trial, MRD assessment using the EAC RT-qPCR assay was found to provide the most powerful independent predictor of disease relapse, being far stronger than the presenting WBC, which has been widely used to dictate treatment approach.44 This study showed that PB affords a reduced sensitivity for MRD detection as compared with BM (median 1.5 log lower sensitivity). Therefore, marrow is the recommended sample source for serial MRD monitoring where the goal is to detect recurrent disease promptly—allowing a sufficient window of time to confirm PCR positivity in an independent sample and to initiate preemptive therapy to prevent progression to frank relapse with its associated risk of fatal bleeding.42 In patients receiving arsenic trioxide (ATO) as salvage therapy, use of MRD monitoring to guide early intervention has been shown to reduce the risk of induction of hyperleukocytosis and the associated differentiation syndrome as compared with treatment in the context of frank relapse.44
Based on the typical sensitivity of RT-qPCR assays for detection of PML-RARA transcripts (∼1 in 104) and kinetics of relapse (median ∼1-log increase in PML-RARA transcripts per month, Figure 4C), BM assessments every 3 months were recommended in the European LeukemiaNet (ELN) guideline.42 MRD monitoring has also been recommended in the National Comprehensive Cancer Network guidelines to inform treatment approach.6 It is important to confirm that patients have achieved molecular remission after frontline therapy in a BM sample affording a sensitivity of at least 1 in 104. However, with survival rates now exceeding 80% among patients treated in clinical trials, particularly those including ATO as frontline therapy, the value of routine sequential monitoring for PML-RARA transcripts beyond the postconsolidation time point has been increasingly questioned.46 Indeed, for patients with low-risk APL (presenting WBC <10 × 109/L) who rapidly achieve molecular remission, based on current evidence there appears to be limited benefit for sequential MRD monitoring beyond the end of treatment (reviewed in Grimwade et al46 ). On the other hand, there remains a case for stringent assessment of MRD in the subgroup of patients presenting with high-risk disease (WBC >10 × 109/L), who have a significant risk of relapse (∼25%) after conventional all-trans retinoic acid and anthracycline-based therapy and can benefit from serial molecular monitoring to guide early salvage with ATO.44 MRD assessment also remains important to guide management of relapsed APL irrespective of WBC at initial presentation (reviewed in Sanz et al42 ).
RUNX1-RUNX1T1 and CBFB-MYH11 detection in CBF leukemias.
Established EAC RT-qPCR assays have also been evaluated in large cohorts of clinical trial patients with core-binding factor (CBF) leukemia, showing that they provide independent prognostic information.47-49 Assessment at early time points during therapy can distinguish patients at significantly differing risk of relapse based on response kinetics.47-50 Importantly, a recent multicenter study conducted by the ALFA and GOELAMS group in 198 patients with CBF leukemia (RUNX1-RUNX1T1 and CBFB-MYH11) showed that MRD response provided more powerful prognostic information in multivariate analysis than results of diagnostic screening for FLT3-ITD and cKIT mutation, with patients failing to achieve a 3-log reduction in leukemic transcript level after 2 courses of chemotherapy being at significantly increased risk of subsequent relapse.49 These findings were extended by Zhu and colleagues, who reported that the poor outcome of patients who (1) failed to achieve a 3-log reduction in RUNX1-RUNX1T1 transcripts compared with a reference pretreatment baseline level (ie, MRD < 0.4%) by the end of the second consolidation or (2) developed early molecular relapse (within 6 months) could be improved by allogeneic transplantation in first complete remission.50 Although the latter study highlights the potential benefit of RT-qPCR for development of risk-directed therapy, several groups have investigated longitudinal MRD monitoring using the standardized EAC assays as a tool to distinguish more precisely those CBF leukemia patients destined to relapse from those who can be cured with chemotherapy alone. Clinically relevant threshold transcript levels were defined, with studies consistently showing that relapse can be predicted by persistently high MRD levels or by a rising trend in transcripts after an initial molecular response.
NPM1 mutant AML.
Frameshift mutations in exon 12 of the NPM1 gene are found in approximately one-third of AML cases, including at least 50% with normal karyotype.1 Although these mutations are heterogeneous (>50 reported to date), they provide an ideal leukemia-specific target for MRD detection by quantitative PCR, using a mutation-specific primer in conjunction with a common primer and probe based on the assay design by Gorello and coworkers.51 Mutation types A, B, and D account for ∼90% of cases for which published primers are available. Mutation-specific primers can be readily designed to allow MRD detection in AML patients with rarer mutations, in which case assays should be carefully tested in control samples (eg, NPM1 wild-type AML) to exclude nonspecific background amplification from the wild-type NPM1 allele. The NPM1 mutant transcript is typically highly expressed in diagnostic AML samples, affording sensitivities typically higher (median 1 in 105) than observed with RT-qPCR assays for other molecular subtypes of AML, with RNA-based assays associated with higher sensitivity than use of genomic DNA. In accordance with the findings in CBF leukemia, RT-qPCR assessment of MRD can distinguish patients at markedly differing risk of relapse based on response kinetics.52-54 Furthermore, relapse can be predicted in individual patients based on persistent high level PCR positivity after frontline therapy or by a rising NPM1 mutant transcript level after an initial molecular response (Figure 4D). The recent study by Shayegi and colleagues also highlighted the potential of serial MRD monitoring to predict outcome after allogeneic transplantation, which could be of value to inform immunosuppression and administration of donor lymphocyte infusion.54
Wilms tumor gene (WT1) expression.
Considering that a significant proportion of AML cases lack an informative leukemia-specific target (ie, chimeric fusion gene, NPM1 mutation, Figure 3), there has been interest as to whether WT1, which is overexpressed in the majority of AML cases, could provide a universal molecular MRD marker. However, there has been inconsistency in the literature concerning the utility of this approach to MRD assessment, which may be a reflection of variations in performance of the numerous published assays, many of which target regions of the gene that are disrupted by mutations.26 This issue has been addressed by an ELN study that systematically evaluated 9 WT1 RT-qPCR assays, leading to selection of an assay that amplified a region outside the mutational hot spots and exhibited the best performance profile.55 A major factor affecting assay sensitivity and clinical utility is that expression of WT1 is not leukemia-specific, with the ELN study establishing an upper limit of expression of 50 and 250 copies/104ABL copies in normal PB and BM, respectively.55 This phenomenon limits the capacity to distinguish low-level MRD from normal background. In contrast to leukemia-specific markers (eg, PML-RARA, NPM1 mutation) in which BM generally provides a more sensitive and reliable sample source for MRD assessment (Figure 4D), in the case of WT1 PB is more informative because of the much higher background level of expression in normal marrow. Taking this into account, based on the analysis of a large cohort of diagnostic AML samples (n = 620), the ELN study established that WT1 is sufficiently highly expressed to allow at least a 2-log reduction in transcript level to be measured in ∼45% of cases. Measurement of kinetics of WT1 response after induction therapy in informative patients can provide independent prognostic information.55 In addition, measurement of WT1 transcript levels has been used in the posttransplant setting to guide the use of donor lymphocyte infusion.56 However, because of the relatively limited sensitivity and lack of specificity of WT1 assessment, this platform seems unlikely to be widely adopted into routine clinical practice, particularly because flow cytometry (see previous sections) and, potentially, newer sequencing-based approaches (see the following section) are expected to be more informative in terms of applicability and sensitivity in the substantial proportion of AML patients in whom MRD tracking is not feasible using an established leukemia-specific RT-qPCR assay.
Newer molecular technologies for MRD detection
Defining the mutational landscape by high-throughput sequencing of AML genomes has broadened the scope of potential molecular methods for MRD detection. Although it is possible to design quantitative PCR assays in the case of recurrent mutations, as exemplified by use of qPCR assays to track MRD posttransplant in JAK2-V617F–associated myeloid neoplasms,57 for point mutations this is challenging because of problems with background amplification from the normal allele. Moreover, because of the marked heterogeneity of mutations already described in AML, developing a catalog of standardized assays to cover every patient would be completely unrealistic. Therefore, several groups have started to explore the use of next-generation sequencing (NGS) technologies as a further platform to detect MRD. To provide proof of principle, Heuser and colleagues used targeted sequencing to successfully detect FLT3-ITD and NPM1 mutations in remission samples and to track the emergence of relapsing disease.58 This work was extended by Kohlmann et al, who highlighted the potential of NGS-based approaches to enable detection of subclinical disease in subsets of AML that are not informative for one of the established leukemia-specific RT-qPCR assays.59 Focusing on AML with RUNX1 mutations, it was shown that with read depths achieving a sensitivity of ∼1 in 1 000, patient outcome could be predicted based on mutational burden at first follow-up. Because this approach is scalable, with increasing read depth, it may be possible to achieve further improvements in sensitivity. However, there are several other technical issues that need to be taken into consideration in the application of NGS for MRD detection, including background sequence error rate and the potential for carryover between sequencing runs, which varies according to protocol and platform.
Digital PCR is another highly promising approach to detect mutations that merits further investigation. This involves partitioning of sample DNA within the PCR mixture using droplet generation or nanofluidic chips depending on the platform, allowing many individual PCRs to be conducted in parallel.60 The partitioning step leads to distribution of single or multiple copies of wild-type, mutant allele, wild-type plus mutant, or no target DNA to each reaction, which are amplified by PCR using different fluorescent probes to distinguish wild-type and mutant copies. The fluorescent readout of each well/droplet is measured individually, allowing precise calculation of the percentage of mutant allele copies in the original sample. This methodology is capable of achieving sensitivities comparable to quantitative PCR, with enhanced capability to distinguish single base mutations from normal background sequence.60 Moreover, there are preliminary data indicating that it may improve MRD detection compared with established standardized RT-qPCR assays in chronic myeloid leukemia.61
Concluding remarks
Discussions with respect to MRD assessment in AML often seem polarized, with questions raised as to whether the information provided (1) adds anything to what is already known about a patient’s prognosis based on conventional risk factors, increasingly complemented by molecular profiling data; and (2) can realistically be used to personalize management and improve clinical outcome, particularly because there is uncertainty about the best course of action to take for any given MRD result. These aspects are being investigated in the UK National Cancer Research Institute AML17/AML19 trials, assessing impact of MRD-directed therapy on outcome, health economics, and quality of life through a “monitor” vs “no monitor” randomization. At present, decisions concerning which particular AML patients should receive transplants can at times seem arbitrary, with clinicians sometimes advocating different strategies in the face of the same information. Based on current evidence, it seems likely that MRD assessment will be useful for making more informed decisions concerning transplantation in first remission. The role of MRD assessment in relation to transplant practice merits more extensive investigation, in particular determining whether the level of disease burden pretransplant could be useful in tailoring the conditioning regimen. Moreover, considering the limited sensitivity of conventional chimerism testing and the dismal outcome of frank relapse after allogeneic transplantation, there could be significant benefits to more widespread uptake of MRD surveillance posttransplant to identify residual disease at an early stage to inform immunosuppression and direct donor lymphocyte infusion.
A further polarizing issue in discussions relating to MRD concerns the question, what is the “best” method? However, given the heterogeneity of AML in terms of mutational and immunophenotypic profile, a “one size fits all” approach (eg, as used in chronic myeloid leukemia) is completely unrealistic, with the most appropriate assay depending upon the characteristics of the leukemia and clinical circumstances (Table 1). It is important to appreciate that methodologies differ in terms of sensitivity and the information they provide. MFC-MRD and genomic DNA-based assays (qPCR, NGS, digital PCR) give a direct measure of leukemic cell burden and are therefore well suited to provide an accurate measure of kinetics of response during early phases of treatment. Unlike RT-qPCR, flow cytometric assays have not been validated for tracking reemergence of leukemia posttherapy. There is, however, some evidence that this is feasible, particularly if the flow cytometric approach takes into account the immunophenotypic changes that result from the evolution of resistant clones from heterogeneous leukemia populations.19,62 For cases with a chimeric fusion gene or NPM1 mutation, the established RT-qPCR assays quantify the relative level of leukemic transcript expression and therefore provide an estimate rather than a direct measure of the burden of residual leukemia. However, for informative patients, these leukemia-specific RT-qPCR assays provide the most sensitive approach to detect residual AML cells and are therefore best suited for sequential MRD monitoring to identify cases with persistent PCR positivity or molecular relapse. Although many studies have defined particular threshold transcript levels that are predictive of outcome at various time points, these “cutoff” values should not be regarded as universally applicable because they may be influenced by a number of parameters including the leukemia and housekeeping gene assays used, sample testing schedule, characteristics of the patient population (eg, age structure), treatment regimen, and length of follow-up. However, the important consistent message emerging from the large comprehensive sequential MRD monitoring studies is that relapse can be reliably predicted by (1) persistently high MRD levels after frontline therapy or (2) a rising trend in transcripts after an initial molecular response.
Proposal as to which platforms are now ready for “prime time” according to AML disease subtype and clinical context
| Platform . | Timing of MRD assessment . | Applicability . | Comments . | |
|---|---|---|---|---|
| Early risk stratification . | Sequential MRD monitoring (longitudinal postconsolidation) . | |||
| Flow cytometry | * | † | >90% | Ready to be used for assessing CR status |
| MRD analysis needs to be conducted in expert laboratories with standardization | ||||
| RT-qPCR | ||||
| Fusion genes/NPM1 mutation | Marker-dependent | * | 32-62%‡ | Standardized assays for leukemia-specific targets available and widely validated |
| WT1 expression | § | † | ∼45% | Assays insensitive and not leukemia-specific, but can predict outcome |
| Next-generation sequencing | † | † | To be established | Promising preliminary data |
| Digital PCR | † | † | To be established | Potentially applicable to detect mutations and fusion genes |
| Platform . | Timing of MRD assessment . | Applicability . | Comments . | |
|---|---|---|---|---|
| Early risk stratification . | Sequential MRD monitoring (longitudinal postconsolidation) . | |||
| Flow cytometry | * | † | >90% | Ready to be used for assessing CR status |
| MRD analysis needs to be conducted in expert laboratories with standardization | ||||
| RT-qPCR | ||||
| Fusion genes/NPM1 mutation | Marker-dependent | * | 32-62%‡ | Standardized assays for leukemia-specific targets available and widely validated |
| WT1 expression | § | † | ∼45% | Assays insensitive and not leukemia-specific, but can predict outcome |
| Next-generation sequencing | † | † | To be established | Promising preliminary data |
| Digital PCR | † | † | To be established | Potentially applicable to detect mutations and fusion genes |
Validated in large studies within clinical trials.
Investigational.
Proportion of AML cases informative for a leukemia-specific molecular marker suitable for detection by RT-qPCR (ie, fusion gene, NPM1 mutation) varies according to age (see Figure 3).
Some supportive data, but not ready for “prime time.”
Improved understanding of the clonal architecture of AML also carries important implications for MRD monitoring strategies. It is now apparent from the examination of sequential samples and paired diagnostic and relapse material that mechanisms underlying clinical relapses are actually quite heterogeneous. Relapses not only may simply reflect reemergence or evolution of the original dominant clone, but can also arise from minor subclones, develop as a second leukemia on the background of the same preleukemic clone (eg, marked by mutations in DNMT3A, IDH, TET2), or represent a completely separate therapy-related leukemia.22-24 These phenomena underlie the “phenotypic shifts” that were highlighted in early immunophenotyping studies and, with respect to molecular-based MRD approaches, emphasize the importance of selecting targets that are considered to be initiating events in leukemogenesis, such as the chimeric fusion genes generated by chromosomal rearrangements (with the caveat that these assays cannot identify emergent therapy-related leukemias). Molecular abnormalities that arise as secondary mutations (eg, FLT3-ITD) may occur in subclones and exhibit less stability over the disease course, thereby limiting their utility as targets for serial MRD monitoring to reliably predict impending relapse. Although acquisition of the NPM1 mutation is not always an initiating event in AML, evidence to date indicates that it nevertheless provides a stable marker of the leukemic clone in the majority of cases.52,53 With more widespread introduction of targeted sequencing into the diagnostic workup of AML to define the mutational profile, it becomes theoretically possible to track MRD in every patient using RT-qPCR or newer DNA-based technologies. Apart from demands on bioinformatics support and greater capacity for data storage, a major challenge will be to establish which mutational targets reliably track the leukemic clone and are most informative to help guide patient management. Achieving standardization of each methodology is important and needs to be accompanied by external quality assurance with clinically relevant QC materials. However, because of the marked heterogeneity of AML, it is clear that laboratories cannot be solely reliant on standard commercial assays, requiring a more flexible approach to fully realize the goal of personalized medicine for every patient with AML.
This article was selected by the Blood and Hematology 2014 American Society of Hematology Education Program editors for concurrent submission to Blood and Hematology 2014. It is reprinted in Hematology Am Soc Hematol Educ Program. 2014;2014:222-233.
Acknowledgments
We would like to thank Nigel Russell, Alan Burnett, Robert Hills, and members of the NCRI AML Working Group for support and enabling evaluation of minimal residual disease in the NCRI AML trials. We thank Adam Ivey and Jelena Jovanovic for data analysis and assistance with preparation of the figures, and Richard Dillon, Paul Virgo, Paresh Vyas, and Krzysztof Mrózek for helpful discussions. We acknowledge Yvonne Morgan, Jennie Lok, Guillermina Nickless, Khalid Tobal, Richard Hall, Beverley Hunt, and the staff of the Molecular Oncology Unit, Guy’s Hospital, London. We also acknowledge Pam Drysdale, Nithiya Clark, Peter Richardson, Steve Dix, Naeem Khan, Tim Plant and staff of the Clinical Immunology Laboratory, University of Birmingham.
This work was supported by the National Institute for Health Research under its Programme Grants for Applied Research Programme (grant RP-PG-0108-10093), Leukaemia & Lymphoma Research of Great Britain, the Guy’s and St. Thomas’ Charity, and the MRD Workpackage (WP12) of the European LeukemiaNet (D.G.).
The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health.
Authorship
Contribution: D.G. and S.D.F. wrote the paper.
Conflict-of-interest disclosure: D.G. declares attendance of an advisory board for BD Biosciences.
Correspondence: David Grimwade, Cancer Genetics Lab, Department of Medical & Molecular Genetics, 8th Floor, Tower Wing, Guy’s Hospital, London SE1 9RT, UK; e-mail: david.grimwade@genetics.kcl.ac.uk.

![Figure 1. Outline of antibody groups used in panels for identification of AML-aberrant immunophenotypes (both for LAIP and “different-from-normal” approaches) and subsequent residual disease monitoring. Core markers are those selected for the backbone of the panel to identify myeloid blast populations. These are combined with markers from lymphoid/myelomonocytic maturation groups (megakaryocytic markers and NG2 [for MLL-rearranged AML] are not included but are more useful in pediatric AML). A stem cell combination may be included (as in the United Kingdom [UK] National Cancer Research Institute [NCRI] AML trial panel) to detect potential immunophenotypic LSC. Most sensitive, robust, aberrant phenotypes include cross-lineage expression and CD34+ human leukocyte antigen (HLA) DR weak/negative. Some aberrant phenotypes may be sensitive (by testing detection by serial dilution in normal marrow) but less stable/useful in follow-up samples. Blue, antibody marker; bold blue, marker in NCRI AML trial panel; red, type of aberrant phenotype potentially detected by marker group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/23/10.1182_blood-2014-05-577593/4/m_3345f1.jpeg?Expires=1769146378&Signature=4~wpW8XR0zAh6WsEIU3yOSh-V08Xz7ur9AL5lM~7zAaNIPhtWtIQCfcvrQbPtJ-jQKFXgYRmNHdfNvHC0dLhWInooHvCzTZDLaat3Z0nlJ~dF4eza7ZYNB3ol~Iit9S8jrvoztt9enRo0jXXme1kCmMXv51rEKSnmsFZk9Z~IOOk4ta6kG8Q0C2QPZwd~AeKo95Cp25WKRjsyc7mVvJ0v-7sF1nICnNlg44Tl2~UtnXQ-PaePPZI4JoInkB7ZpoTkF13l1k7xMeDrc~kfm6OqOeOK8VF86i2KdZiYijBL9telhEFtSmB6KmO2x7uPQF4jUfYXmfXNPQmntiMukti5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)