Key Points
GPR84 simultaneously augments β-catenin signaling and an oncogenic transcription program essential for establishment of MLL.
Our study demonstrates a strong dependence of hematopoietic stem cell–derived MLL leukemic cells on GPR84 for disease maintenance in vivo.
Abstract
β-catenin is required for establishment of leukemic stem cells (LSCs) in acute myeloid leukemia (AML). Targeted inhibition of β-catenin signaling has been hampered by the lack of pathway components amenable to pharmacologic manipulation. Here we identified a novel β-catenin regulator, GPR84, a member of the G protein-coupled receptor family that represents a highly tractable class of drug targets. High GPR84 expression levels were confirmed in human and mouse AML LSCs compared with hematopoietic stem cells (HSCs). Suppression of GPR84 significantly inhibited cell growth by inducing G1-phase cell-cycle arrest in pre-LSCs, reduced LSC frequency, and impaired reconstitution of stem cell–derived mixed-lineage leukemia (MLL) AML, which represents an aggressive and drug-resistant subtype of AML. The GPR84-deficient phenotype in established AML could be rescued by expression of constitutively active β-catenin. Furthermore, GPR84 conferred a growth advantage to Hoxa9/Meis1a-transduced stem cells. Microarray analysis demonstrated that GPR84 significantly upregulated a small set of MLL-fusion targets and β-catenin coeffectors, and downregulated a hematopoietic cell-cycle inhibitor. Altogether, our data reveal a previously unrecognized role of GPR84 in maintaining fully developed AML by sustaining aberrant β-catenin signaling in LSCs, and suggest that targeting the oncogenic GPR84/β-catenin signaling axis may represent a novel therapeutic strategy for AML.
Introduction
The ability for self-renewal, migration/invasion, and drug resistance are fundamental properties of malignant stem cells that drive both disease progression and relapse. Identification of pathways and their molecular components essential for the regulation of abnormally acquired stem cell–like properties is a prerequisite for understanding the underlying mechanisms of oncogenesis and designing effective anticancer therapeutic strategies. We and others have previously shown that mixed-lineage leukemia (MLL) fusion proteins can aberrantly activate Wnt/β-catenin signaling in hematopoietic stem cells (HSCs) or more differentiated granulocyte–macrophage progenitors (GMPs) for establishment of leukemic stem cells (LSCs) in acute myeloid leukemia subtype M5 (AML-M5)1,2 ; however, the mechanisms involved remain obscure. It has also been noted that activation of β-catenin is observed in tumors without clear mutations in major components of this pathway or an increase in Wnt signaling.3 This suggests that other developmental signaling pathways may be capable of inducing activation or downstream signaling of β-catenin.
G protein–coupled receptors (GPCRs), the largest family of cell-surface molecules with key roles in transmitting signals to downstream effectors, have emerged as crucial players in promoting tumor growth and metastasis1,4-8 and represent one of the most important drug targets in pharmaceutical development.9,10 Recent evidence indicates that GPCRs are active in various cancer cell lines and primary patient samples,7,8 and that oncogenic GPCRs augment activation of β-catenin signaling in tumor cells.4,7,11 The importance of GPCRs in regulation of β-catenin signaling and tumorigenesis has been increasingly recognized.4,5,7,9 A recent report has identified GPR84 (a proinflammatory GPCR)12 as an LSC-specific gene with its expression level significantly higher in human AML LSCs than that in HSCs in a genome-wide gene-expression analysis and subsequent quantitative real-time reverse-transcriptase polymerase chain reaction (RT-PCR).13 Interestingly, the high level of GPR84 expression is also observed exclusively in human AML-M5 LSCs compared with other LSCs and HSCs,14 indicating a possible disease subtype–dependent expression. This finding is in line with substantially higher levels of GPR84 expression found in human AML-M5 cells carrying the MLL-AF9 translocation (ie, THP1) than in human myeloid cell lines from other subtypes including Kasumi-3, K051/K052, HL-60, HNT-34, and UCSD/AML1.15 In addition, lipopolysaccharide, a potent inducer of inflammation, significantly elevates GPR84 expression in THP1,16 suggesting that an inflammatory mechanism is potentially involved. However, it remains unclear whether this receptor is functionally involved in leukemogenesis. In this study, microarray analysis has confirmed the upregulation of GPR84 expression in mouse MLL LSCs compared with normal stem cells. Using the mouse MLL-AF9–driven AML model that shares common features with human MLL AML,17,18 we explored GPR84-mediated activation of β-catenin signaling and its functional significance in leukemia maintenance of a poor-prognosis AML subtype that is highly resistant to current chemotherapy.19,20 Our results provide the first evidence for an essential role of GPR84 in stem cell–derived MLL leukemia.
Methods
See supplemental Data for additional methods (available on the Blood Web site).
Mice
Six- to 8-week-old C57BL/6 mice (Australian BioResources, Mossvale, NSW, Australia) were used as bone marrow (BM) donors and recipients. All animal studies were approved by the Animal Care and Ethics Committee of the University of New South Wales.
Hematopoietic cell isolation and viral transduction
HSC-enriched KLS were sorted as Lin– (CD3, CD4, CD8a, CD19, B220, Gr-1, Ter119, IL-7R) Sca-1+ c-kit+ (BioLegend, San Diego, CA), and myeloid progenitor GMPs were isolated as Lin− Sca-1− c-kithi CD34+ FcγRII/IIIhi (BioLegend) from mouse BM as previously described.1,17,21 For viral transduction, 1-5 × 104 cells were incubated with concentrated virus supplemented with 7.5 µg/mL polybrene (Sigma-Aldrich/Merck, Castle Hill, NSW, Australia) and cytokines including 10 ng/mL mIL-3, 10 ng/mL mIL-6, and 25 ng/mL mSCF (StemCell Technologies, Tullamarine, VIC, Australia). Two rounds of transduction were performed for each experiment.
Colony formation
GFP+ pre-LSCs or leukemic cells were plated (1000 cells per 35-mm dish) in M3234 methylcellulose (StemCell Technologies) supplemented with cytokines. Colonies were counted after 5 days, and cells were subsequently replated onto secondary methylcellulose dishes. Three rounds of replating were performed for each experiment.
Microarray data analysis
Total RNA was extracted as described in the supplemental Methods and analyzed using Illumina mouse WG-6 v2.0 expression arrays. Microarray data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1932. Labeling, hybridization, and scanning were performed by the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Sydney, NSW, Australia), with each sample hybridized to an individual array. Microarray results were analyzed using GenePattern software (Broad Institute, Massachusetts Institute of Technology) as previously described.1,17 Significantly enriched gene sets were determined by the Gene Set Enrichment Analysis (GSEA) method (http://www.broadinstitute.org/gsea).22
Results
GPR84 is aberrantly upregulated in human and mouse MLL AML
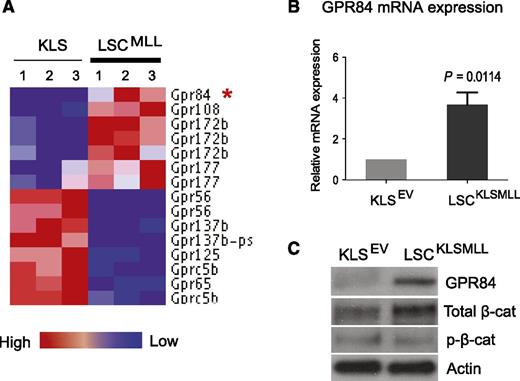
Previous reports demonstrated aberrant upregulation of GPR84 expression in human primary AML LSCs.13 To determine whether MLL-fusion oncoproteins can alter expression of GPCR genes, we analyzed gene expression profiles of GPCRs in mouse and human MLL–rearranged AML. We previously demonstrated that a Lin− Sca-1− c-kithi CD34+ FcγRII/III+ subpopulation is enriched for LSCs (also known as LGMP or GMP-like LSCs) in the MLL-AF9 AML mouse model,17 and we conducted a DNA microarray analysis to compare genes differentially expressed between mouse MLL LSCs and HSC-enriched KLS cells (GEO #GSE20377).1 Here, we generated a GPCR gene set with defined expression levels using our microarray data and found that GPR84 along with GPR108, GPR172b, and GPR177 were upregulated, whereas 6 GPCRs were downregulated by MLL-AF9 in mouse LSCs compared with KLS cells (Figure 1A). Expression of these upregulated receptors was further analyzed using a published gene expression data set comparing MLL-rearranged human primary patient samples with normal human HSCs,23 and the results revealed a greater than eightfold increase in GPR84 expression compared with a 1.8- to 2.4-fold increase in 3 other GPCR genes in human MLL AML (supplemental Figure 1). These findings suggest potential clinical relevance for upregulation of GPR84 in MLL AML.
GPR84 is upregulated in mouse MLL LSCs, and there is a positive correlation between GPR84 and β-catenin expression. (A) The heat map of the 10 GPCR genes, which passed a fold-change cutoff of 1.5. Expression of GPR84 in MLL LSCs compared with HSC-enriched KLS cells is marked with an red asterisk. (B) Real-time RT-PCR confirmed the microarray result with primers specific for GPR84 in KLS cells transduced with empty vector (EV) and KLSMLL LSCs isolated from mice with an established MLL-AF9 AML initiated from KLS cells. The results were normalized using GAPDH as a control. (C) Western blot analysis confirmed changes in the protein expression of GPR84 and β-catenin (total or inactive/phosphorylated β-catenin) in KLS cells expressing EV and KLSMLL LSCs.
GPR84 is upregulated in mouse MLL LSCs, and there is a positive correlation between GPR84 and β-catenin expression. (A) The heat map of the 10 GPCR genes, which passed a fold-change cutoff of 1.5. Expression of GPR84 in MLL LSCs compared with HSC-enriched KLS cells is marked with an red asterisk. (B) Real-time RT-PCR confirmed the microarray result with primers specific for GPR84 in KLS cells transduced with empty vector (EV) and KLSMLL LSCs isolated from mice with an established MLL-AF9 AML initiated from KLS cells. The results were normalized using GAPDH as a control. (C) Western blot analysis confirmed changes in the protein expression of GPR84 and β-catenin (total or inactive/phosphorylated β-catenin) in KLS cells expressing EV and KLSMLL LSCs.
We next examined the differential expression of GPR84 in mouse KLS cells and LSCs isolated from mice with an established MLL-AF9 AML initiated from KLS cells (KLSMLL). Quantitative real-time RT-PCR and western blot analyses confirmed the upregulation of GPR84 by MLL-AF9 at both mRNA and protein levels in LSCs (Figure 1B-C and supplemental Figure 2). Furthermore, western blot analysis revealed an increase in the level of total β-catenin protein, with no change in inactive/phosphorylated β-catenin (p-β-cat; Figure 1C), indicating the activation of β-catenin in LSCs compared with KLS cells. The positive correlation between the expression of GPR84 and active β-catenin in LSCs implicates a potential role for GPR84 in the leukemogenic function of β-catenin.
Knockdown of GPR84 represses β-catenin expression and impairs MLL-AF9–mediated leukemic transformation in vitro
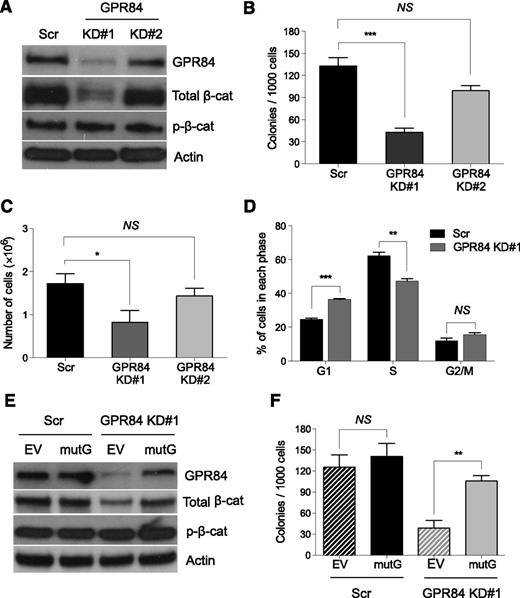
To determine whether GPR84 regulates β-catenin expression, effective knockdown of GPR84 in KLSMLL pre-LSCs, which are defined as KLS cells transduced with MLL-AF9 oncogene that have the ability to induce leukemia upon transplantation into mice with a relatively long latency period,1,19 was achieved by lentiviral-mediated delivery of an shRNA (KD#1) for GPR84. After puromycin selection, GPR84 and β-catenin protein levels in transduced pre-LSCs were analyzed by Western blot analyses. The results showed that KD#1-mediated stable knockdown of GPR84 markedly decreased the expression of endogenous active β-catenin (Figure 2A), suggesting that GPR84 functions as a positive regulator of β-catenin protein expression.
GPR84 knockdown suppresses β-catenin expression and in vitro colony formation of stem cell–derived MLL pre-LSCs. (A) Western blot analysis of GPR84 knockdown efficiency and β-catenin expression in scrambled control (Scr) or 2 independent GPR84 shRNAs (KD#1 and KD#2) transduced KLSMLL pre-LSCs. (B) Colony-forming assays of Scr or GPR84 KD KLSMLL pre-LSCs. Numbers of colonies per dish at the third round of replating are shown. (C) Total number of cells per dish after the third round of replating. (D) Flow cytometric analysis of cell-cycle status in Scr or GPR84 KD#1 KLSMLL pre-LSCs. Error bars represent triplicates obtained in independent experiments. (E) Western blots of GPR84 and β-catenin in Scr or GPR84 KD#1 KLSMLL pre-LSCs expressing empty vector (EV) or KD#1-resistant GPR84 cDNA (mutG). (F) Colony-forming assays of Scr or GPR84 KD#1 KLSMLL pre-LSCs expressing EV or mutG. *P < .05, **P < .01, ***P < .0005, NS, not significant (P > .05).
GPR84 knockdown suppresses β-catenin expression and in vitro colony formation of stem cell–derived MLL pre-LSCs. (A) Western blot analysis of GPR84 knockdown efficiency and β-catenin expression in scrambled control (Scr) or 2 independent GPR84 shRNAs (KD#1 and KD#2) transduced KLSMLL pre-LSCs. (B) Colony-forming assays of Scr or GPR84 KD KLSMLL pre-LSCs. Numbers of colonies per dish at the third round of replating are shown. (C) Total number of cells per dish after the third round of replating. (D) Flow cytometric analysis of cell-cycle status in Scr or GPR84 KD#1 KLSMLL pre-LSCs. Error bars represent triplicates obtained in independent experiments. (E) Western blots of GPR84 and β-catenin in Scr or GPR84 KD#1 KLSMLL pre-LSCs expressing empty vector (EV) or KD#1-resistant GPR84 cDNA (mutG). (F) Colony-forming assays of Scr or GPR84 KD#1 KLSMLL pre-LSCs expressing EV or mutG. *P < .05, **P < .01, ***P < .0005, NS, not significant (P > .05).
To dissect the role of GPR84 in MLL-AF9 leukemic transformation, we performed methylcellulose replating assays. The results showed that GPR84 knockdown resulted in a significant reduction in colony formation and proliferative capacity of pre-LSCs (Figure 2B-C). This suggests that KD#1 is sufficient to induce a GPR84-deficient phenotype in vitro. Furthermore, flow cytometric analysis showed that, compared with control cells expressing a scrambled shRNA (Scr), GPR84 KD cells revealed a high percentage of cells in the G1 phase and a low percentage of cells in the S phase (Figure 2D), indicating a defect of G1-to-S phase cell-cycle transition. To confirm the specificity of shRNA-mediated silencing of GPR84, KD#1 and Scr pre-LSCs were transduced with a retrovirus-encoding empty vector (EV) or a mutant GPR84 cDNA (mutG) that is not targeted by the GPR84 shRNA, respectively. The mutated cDNA sequence (CtGTcGAtACtTAttTgCA) does not alter the amino-acid sequence of GPR84. As expected, forced overexpression of mutG not only resulted in the reactivation of β-catenin in GPR84 KD pre-LSCs (Figure 2E) but also rescued the GPR84-deficient phenotype (Figure 2F). Together, these data suggest that GPR84 is essential for MLL-AF9 leukemic transformation in vitro through promotion of β-catenin signaling.
GPR84 is required for maintenance of established MLL
To evaluate the leukemogenic function of GPR84 in vivo, we first studied its role in AML development. Genetically modified GFP+ KLSMLL pre-LSCs were transplanted into sublethally irradiated primary recipient mice. Given the impaired growth observed in GPR84 KD pre-LSCs in vitro, the results unexpectedly revealed no effect of GPR84 loss on the survival of primary recipients (Figure 3A). This is likely caused by the incomplete knockdown of GPR84 in pre-LSCs, although the knockdown level was clearly sufficient to compromise in vitro transformation. BM of leukemic mice that received GPR84 KD and Scr pre-LSCs were typically replaced by GFP+ leukemic cells to a level of ∼85% and 95%, respectively (Figure 3B). However, it was noted that GPR84 KD pre-LSCs showed reduced infiltration into the spleen as manifested by a relatively low percentage of GFP+ cells (Figure 3B). As such, spleen weights were significantly lower in mice transplanted with GPR84 KD pre-LSCs than in control mice (Figure 3C). Flow cytometric analysis confirmed persistent GPR84 knockdown and suppression of endogenous β-catenin protein expression in GFP+ leukemic cells from primary recipients (Figure 3D-E). This suggests that knockdown of GPR84 is sufficient to block dissemination of leukemic cells to secondary organs.
GPR84 knockdown blocks dissemination of leukemic cells to the spleen and compromises leukemia reconstitution. (A) Kaplan-Meier survival curves for mice injected with 1 × 106 Scr or GPR84 KD#1 KLSMLL GFP+ pre-LSCs. (B) Percentage of GFP+ leukemic cells in BM and spleen. NS, not significant (P > .05). (C) Spleen weights of primary AML mice. (D-E) Flow cytometric analysis of GPR84 and β-catenin protein levels in GFP+ leukemic cells from primary recipients. (F-G) Kaplan-Meier survival curves for mice that received 1 × 105 GFP+ leukemic cells from primary recipients in secondary transplantation assays, or that received the indicated number (102-104) of GFP+ leukemic cells for in vivo limiting dilution assessment. Calculated LSC frequencies in GFP+ Scr and GPR84 KD#1 leukemic cells are 1 in 433 and 1 in 1254, respectively. (H-I) Representative histograms illustrating in vivo BrdU incorporation cell proliferation assays of GFP+ leukemic cells from the BM of secondary recipients 10 days after transplantation. (J) Bar graphs representing quantitative analysis of the data obtained from in vivo BrdU incorporation assays (indicative of 6 independent experiments).
GPR84 knockdown blocks dissemination of leukemic cells to the spleen and compromises leukemia reconstitution. (A) Kaplan-Meier survival curves for mice injected with 1 × 106 Scr or GPR84 KD#1 KLSMLL GFP+ pre-LSCs. (B) Percentage of GFP+ leukemic cells in BM and spleen. NS, not significant (P > .05). (C) Spleen weights of primary AML mice. (D-E) Flow cytometric analysis of GPR84 and β-catenin protein levels in GFP+ leukemic cells from primary recipients. (F-G) Kaplan-Meier survival curves for mice that received 1 × 105 GFP+ leukemic cells from primary recipients in secondary transplantation assays, or that received the indicated number (102-104) of GFP+ leukemic cells for in vivo limiting dilution assessment. Calculated LSC frequencies in GFP+ Scr and GPR84 KD#1 leukemic cells are 1 in 433 and 1 in 1254, respectively. (H-I) Representative histograms illustrating in vivo BrdU incorporation cell proliferation assays of GFP+ leukemic cells from the BM of secondary recipients 10 days after transplantation. (J) Bar graphs representing quantitative analysis of the data obtained from in vivo BrdU incorporation assays (indicative of 6 independent experiments).
Impaired infiltration of GPR84 KD KLSMLL pre-LSCs into the spleen prompted us to assess the effect of GPR84 knockdown on the maintenance of established leukemia. Sorted GFP+ leukemic cells from primary recipients were transplanted into secondary recipients. In contrast to primary leukemia, there was a significant survival advantage in the GPR84 KD mice, and knockdown of GPR84 impaired reconstitution of MLL leukemia and significantly prolonged the survival of secondary recipients (Figure 3F). In vivo limiting dilution transplantation assays showed that GPR84 knockdown reduced the frequency of AML LSCs (Figure 3G). Further in vivo BrdU labeling analysis revealed an evident decrease in proliferating BrdU+ cells in the BM of secondary GPR84 KD recipient mice (Figure 3H-J), indicating a defect in leukemic cell proliferation in vivo. Collectively, these results suggest that GPR84 knockdown impairs LSC function and proliferation of established leukemia in vivo.
We and others have previously demonstrated that pre-LSCs of KLSMLL and Hoxa9/Meis1a-transduced KLS cells (KLSA9M) have a similar immunophenotype in methylcellulose and produce histopathologically similar diseases1,17,19 ; yet KLSA9M cells express a lower level of β-catenin (supplemental Figure 3) and develop a less aggressive AML with prolonged survival in mice.1,19 This provides us with an assay system to examine the effect of GPR84 knockdown on in vivo leukemogenic phenotypes of pre-LSCs expressing different levels of β-catenin. We transplanted GPR84 KD or Scr KLSA9M pre-LSCs into primary recipient mice. Intriguingly, mice receiving GPR84 KD cells showed a significantly longer survival, with 50% still healthy at the time of sacrifice compared with control mice (supplemental Figure 4). Thus, the KD#1-mediated knockdown of GPR84 is sufficient to impair the development of leukemia initiated by KLSA9M pre-LSCs, which express relatively lower levels of β-catenin than KLSMLL pre-LSCs. These data suggest that loss of GPR84 is a critical determinant of the leukemic phenotype in vivo, with the effect being dependent on the level of β-catenin present. The results further support the important role of GPR84 in AML leukemogenesis.
β-catenin activation rescues GPR84 deficiency in established AML
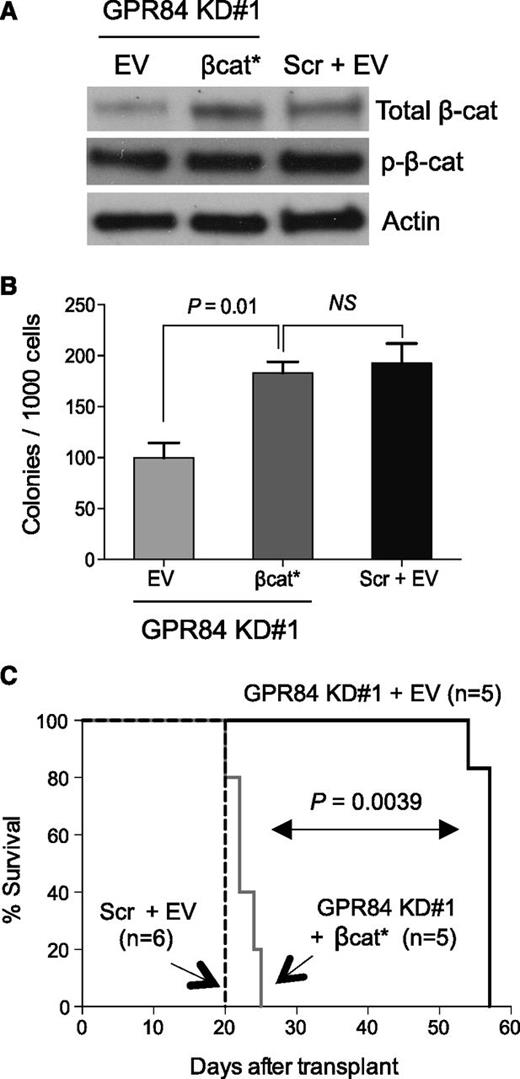
To assess whether β-catenin is functionally regulated by GPR84 and to determine the ability of activated β-catenin to rescue the defective LSC function caused by GPR84 knockdown in established leukemia, sorted GFP+ leukemic cells from primary recipients were transduced with a retrovirus encoding a constitutively active form of β-catenin (βcat*), and subsequent Western blot analysis confirmed enforced βcat* overexpression (Figure 4A). βcat* expression significantly enhanced the colony-forming capacity of GPR84 KD leukemic cells and rescued GPR84 deficiency in vitro (Figure 4B). Furthermore, βcat*-expressing GPR84 KD KLSMLL leukemic cells produced secondary AML with kinetics similar to KLSMLL leukemic cells and with considerably shorter latency compared with GPR84 KD KLSMLL leukemic cells (Figure 4C). This suggests that β-catenin activation itself is capable of rescuing GPR84 deficiency in vivo, and that the block in cell growth and proliferation can be attributed primarily to reduced β-catenin levels. Our data collectively indicate that β-catenin is a functional downstream target of GPR84, and GPR84-mediated β-catenin activation contributes to leukemia maintenance.
Expression of constitutively active β-catenin rescues the GPR84-deficient phenotype in vitro and in vivo. Scr or GPR84 KD#1 KLSMLL GFP+ leukemic cells were sorted from BM cells of primary recipients and subsequently transduced with constitutively active β-catenin (βcat*) or empty vector (EV). (A) Western blot analysis of β-catenin expression in transduced GFP+ KLSMLL leukemic cells. (B) Colony-forming assays of transduced GFP+ KLSMLL leukemic cells. NS, not significant (P > .05). Mean of assay performed in triplicate. (C) Kaplan-Meier survival curves for secondary recipient mice that received 1 × 105 transduced GFP+ KLSMLL leukemic cells.
Expression of constitutively active β-catenin rescues the GPR84-deficient phenotype in vitro and in vivo. Scr or GPR84 KD#1 KLSMLL GFP+ leukemic cells were sorted from BM cells of primary recipients and subsequently transduced with constitutively active β-catenin (βcat*) or empty vector (EV). (A) Western blot analysis of β-catenin expression in transduced GFP+ KLSMLL leukemic cells. (B) Colony-forming assays of transduced GFP+ KLSMLL leukemic cells. NS, not significant (P > .05). Mean of assay performed in triplicate. (C) Kaplan-Meier survival curves for secondary recipient mice that received 1 × 105 transduced GFP+ KLSMLL leukemic cells.
GPR84 confers a growth advantage to pre-LSCs for AML development
Given the evidence that KLSA9M pre-LSCs develop leukemia with a much longer latency compared with KLSMLL pre-LSCs,1,19 and that β-catenin is required for establishment of MLL LSCs,1,2 we hypothesized that GPR84-augmented activation of β-catenin can enhance Hoxa9/Meis1-induced leukemogenesis and likely contributes to the full-blown MLL phenotype. To test this, we transduced KLSA9M pre-LSCs with a retrovirus-encoding GPR84 cDNA or empty vector. Remarkably, enforced overexpression of GPR84 not only substantially activated endogenous β-catenin expression (Figure 5A), further demonstrating a strong positive correlation between GPR84 and β-catenin expression, but also potentiated the clonogenic growth of KLSA9M pre-LSCs in vitro (Figure 5B). When these cells were injected into syngeneic recipient mice, GPR84 overexpression significantly accelerated the onset of KLSA9M-induced AML and developed leukemia with similar latency to that of KLSMLL group (Figure 5C). To characterize the in vivo effect of GPR84 upregulation, we examined the short-term proliferative capacity of GFP+ KLSA9M pre-LSCs in recipient mice. Strikingly, ectopic expression of GPR84 enhanced in vivo cell proliferation, as indicated by a noticeable increase in GFP+ BM cells observed at 10 days post-transplantation (Figure 5D). Thus, GPR84 overexpression confers an apparent growth advantage in vitro and in vivo through augmenting β-catenin signaling and promotes leukemia formation.
GPR84 confers a growth advantage on KLSA9M pre-LSCs. (A) Western blot analysis of GPR84 and β-catenin expression in KLSA9M pre-LSCs transduced with GPR84 cDNA or EV, as well as KLSMLL pre-LSCs transduced with EV. (B) Colony-forming assays of KLSA9M pre-LSCs expressing GPR84 cDNA or EV (N = 3). (C) Kaplan-Meier survival curves for mice that received 1 × 106 KLSMLL pre-LSCs expressing EV, KLSA9M pre-LSCs expressing GPR84 cDNA, or EV. (D) Representative flow cytometry plots illustrating in vivo short-term cell proliferation assays of GFP+ KLSA9M pre-LSCs expressing GPR84 or EV 10 days after transplantation (N = 3 independent experiments). BM cells isolated from normal mice were used as a GFP-negative control to set up an appropriate flow cytometric gate.
GPR84 confers a growth advantage on KLSA9M pre-LSCs. (A) Western blot analysis of GPR84 and β-catenin expression in KLSA9M pre-LSCs transduced with GPR84 cDNA or EV, as well as KLSMLL pre-LSCs transduced with EV. (B) Colony-forming assays of KLSA9M pre-LSCs expressing GPR84 cDNA or EV (N = 3). (C) Kaplan-Meier survival curves for mice that received 1 × 106 KLSMLL pre-LSCs expressing EV, KLSA9M pre-LSCs expressing GPR84 cDNA, or EV. (D) Representative flow cytometry plots illustrating in vivo short-term cell proliferation assays of GFP+ KLSA9M pre-LSCs expressing GPR84 or EV 10 days after transplantation (N = 3 independent experiments). BM cells isolated from normal mice were used as a GFP-negative control to set up an appropriate flow cytometric gate.
GPR84 positively regulates an oncogenic transcription program essential for establishment of MLL leukemia and Wnt/β-catenin–associated gene signatures
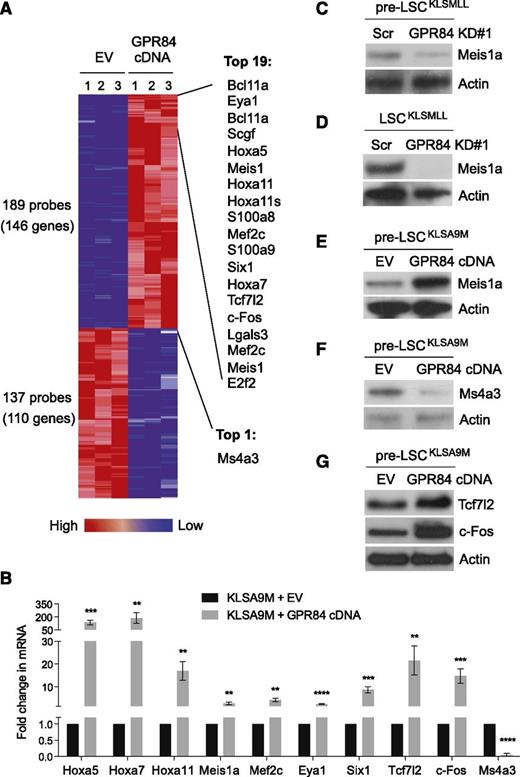
We next examined gene expression changes regulated by GPR84 in KLSA9M pre-LSCs using DNA microarray analysis. A subset of genes (146 genes upregulated, 110 genes downregulated by GPR84) passed the stringent statistical cutoff of P < .03 and fold change ≥3 (Figure 6A). Data from the microarray analysis were then validated by real-time RT-PCR (Figure 6B). The results showed that GPR84 preferentially upregulated a small set of known MLL-fusion target genes including Hox genes (ie, Hoxa5, Hoxa7, Hoxa11), Meis1a, Eya1, Six1, and Mef2c,1,17,24-29 suggesting its role in the regulation of MLL target genes. Western blot analysis further verified GPR84-mediated expression of Meis1a, an essential regulator of MLL LSC potential and frequency,30 in GPR84 KD KLSMLL pre-LSCs (Figure 6C) or LSCs (Figure 6D), and in GPR84-overexpressing KLSA9M pre-LSCs (Figure 6E and supplemental Figure 5). Intriguingly, our real-time RT-PCR and western blot analyses (Figure 6B,F) confirmed that GPR84 overexpression led to a substantial reduction in the expression of a G1-S phase cell-cycle inhibitor, Ms4a3.31,32 This could thus explain the result shown in Figure 2D that GPR84 knockdown induced G1 arrest in KLSMLL pre-LSCs, likely through reactivation of Ms4a3.
GPR84 upregulates a small set of genes essential for establishment of MLL leukemia while activating β-catenin transcriptional cofactors. (A) The heat map displaying the probe sets/genes differentially expressed in GPR84- and EV-expressing KLSA9M pre-LSCs. The ComparativeMarkerSelection module from GenePattern was used to analyze 3 biological replicates from each cell type using a fold change of ≥3 and P < .03 as the cutoff criteria. The top 20 genes up- and downregulated by GPR84 overexpression are listed on the right. (B) Confirmation of microarray results by real time RT-PCR (N = 4) in KLSA9M pre-LSCs expressing GPR84 cDNA or EV. **P < .01, ***P < .0005, ****P < .0001. Western blot analysis of Meis1a expression in GPR84 KD#1 KLSMLL pre-LSCs (C) or LSCs (D), and in GPR84-overexpressing KLSA9M pre-LSCs (E); western blot analysis of Ms4a3 expression in EV or GPR84-overexpressing KLSA9M pre-LSCs (F); western blot analysis of Tcf7l2 and c-Fos expression in EV or GPR84-overexpressing KLSA9M pre-LSCs (G).
GPR84 upregulates a small set of genes essential for establishment of MLL leukemia while activating β-catenin transcriptional cofactors. (A) The heat map displaying the probe sets/genes differentially expressed in GPR84- and EV-expressing KLSA9M pre-LSCs. The ComparativeMarkerSelection module from GenePattern was used to analyze 3 biological replicates from each cell type using a fold change of ≥3 and P < .03 as the cutoff criteria. The top 20 genes up- and downregulated by GPR84 overexpression are listed on the right. (B) Confirmation of microarray results by real time RT-PCR (N = 4) in KLSA9M pre-LSCs expressing GPR84 cDNA or EV. **P < .01, ***P < .0005, ****P < .0001. Western blot analysis of Meis1a expression in GPR84 KD#1 KLSMLL pre-LSCs (C) or LSCs (D), and in GPR84-overexpressing KLSA9M pre-LSCs (E); western blot analysis of Ms4a3 expression in EV or GPR84-overexpressing KLSA9M pre-LSCs (F); western blot analysis of Tcf7l2 and c-Fos expression in EV or GPR84-overexpressing KLSA9M pre-LSCs (G).
We also noted a pronounced increase in both mRNA and protein levels of β-catenin transcriptional cofactors including Tcf7l2 and c-Fos in GPR84-overexpressing KLSA9M pre-LSCs (Figure 6B,G),33-37 indicating GPR84-mediated potentiation of Wnt/β-catenin signaling. Consistently, GSEA22 of our microarray expression data showed that expression of Wnt/β-catenin target genes, which have been previously identified in various normal and tumor tissues38-45 (Figure 7A) and in known Wnt-associated gene sets46 (Figure 7C), was highly enriched among genes upregulated by GPR84 overexpression in KLSA9M pre-LSCs. Real-time RT-PCR confirmed the changes in expression of the top 7 Wnt/β-catenin target genes upregulated by GPR84 overexpression in KLSA9M pre-LSCs (Figure 7B). Together, these data highlight a significant overlap between the genes upregulated by GPR84 and those upregulated by MLL-fusions or in Wnt/β-catenin signaling (supplemental Figure 6), supporting an essential role of GPR84 in MLL transcriptional regulation and leukemogenesis.
GSEA of our array data reveals GPR84-mediated enrichment of gene expression signatures for Wnt/β-catenin signaling. (A) The plot of β-catenin (CTNNB1) target GSEA22 for GPR84-overexpressing KLSA9M pre-LSCs. Normalized gene expression data were used for GSEA (http://www.broad.mit.edu/gsea). The green curve of the plot showing the running enrichment score (ES) for the TARGET_GENES_CTNNB1 gene set, which was generated using Wnt/β-catenin target genes identified in various tumors38-42 and normal tissues43-45 (listed at http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes). The rectangle indicating the “leading edge” subset (a subgroup of the “most upregulated” genes by GPR84 overexpression) in their ranked order. The list on the right shows the top 14 genes in the leading edge subset. (B) Real time RT-PCR (N = 3) confirmed the expression of the top 7 genes in EV or GPR84-overexpressing KLSA9M pre-LSCs. *P < .05, **P < .01, ****P < .0001. (C) Enrichment plots for statistically significant gene sets identified by GSEA from the molecular signature database (MSigDB)46 exhibiting upregulation of Wnt-associated gene signatures upon GPR84 overexpression. False discovery rate (FDR) gives the estimated probability that a gene set with a given normalized ES (NES) represents a false-positive finding; FDR <0.25 is a widely accepted cutoff for the identification of biologically significant gene sets.22
GSEA of our array data reveals GPR84-mediated enrichment of gene expression signatures for Wnt/β-catenin signaling. (A) The plot of β-catenin (CTNNB1) target GSEA22 for GPR84-overexpressing KLSA9M pre-LSCs. Normalized gene expression data were used for GSEA (http://www.broad.mit.edu/gsea). The green curve of the plot showing the running enrichment score (ES) for the TARGET_GENES_CTNNB1 gene set, which was generated using Wnt/β-catenin target genes identified in various tumors38-42 and normal tissues43-45 (listed at http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes). The rectangle indicating the “leading edge” subset (a subgroup of the “most upregulated” genes by GPR84 overexpression) in their ranked order. The list on the right shows the top 14 genes in the leading edge subset. (B) Real time RT-PCR (N = 3) confirmed the expression of the top 7 genes in EV or GPR84-overexpressing KLSA9M pre-LSCs. *P < .05, **P < .01, ****P < .0001. (C) Enrichment plots for statistically significant gene sets identified by GSEA from the molecular signature database (MSigDB)46 exhibiting upregulation of Wnt-associated gene signatures upon GPR84 overexpression. False discovery rate (FDR) gives the estimated probability that a gene set with a given normalized ES (NES) represents a false-positive finding; FDR <0.25 is a widely accepted cutoff for the identification of biologically significant gene sets.22
Discussion
Although the cell of origin has been implicated as a determinant of in vivo response to chemotherapy in MLL AML,19 and as such MLL pre-LSCs that originate from stem cells develop a particular aggressive and drug-resistant subtype of AML,19 very little is known about the molecular mechanisms underlying this important disease subtype. In this study, we provide the first evidence for a crucial role of GPR84 in AML development and maintenance. Because overexpression of GPR84 augments β-catenin signaling and results in significantly elevated expression of genes required for in vivo dissemination and oncogenic properties of MLL leukemic cells, the GPR84/β-catenin signaling axis is likely to represent an attractive yet unexplored therapeutic target in MLL AML.
Our study demonstrates that GPR84 is essential for LSC function and maintenance of established MLL AML in mice, and that GPR84 is upregulated by MLL-AF9 in murine KLS–derived LSCs, which is consistent with aberrant upregulation of GPR84 mRNA expression in human AML LSCs compared with normal HSCs.13,14 We and others have previously demonstrated that the levels of β-catenin expression are critical in determining MLL-fusion–mediated malignant transformation.1,2 Herein, we show that GPR84 positively regulates β-catenin expression at different stages (pre-LSC to LSC) of leukemia, and that the functional defect caused by GPR84 deficiency can be largely rescued in vitro and in vivo by re-expression of active β-catenin. Importantly, GPR84 overexpression not only activates β-catenin transcriptional cofactors Tcf7l2 and c-Fos but also significantly associates with genes upregulated in Wnt/β-catenin signaling, further supporting its involvement in pathway activation during malignant transformation. This is in agreement with previous observations that aberrant activation of β-catenin is required for self-renewal of MLL LSCs,1,2 whereas suppression of β-catenin impairs LSC function and significantly reduces the growth of human MLL leukemic cells.1,2 These findings highlight a novel role for GPR84 in the β-catenin signaling cascade in MLL AML.
Consistently, emerging evidence shows that GPCRs can exert their tumorigenic functions through modulating β-catenin signaling in cancers, including colon cancer.1,4-7 Although 2 GPCRs, prostaglandin E receptor 2 (EP2) and GPR48, have been respectively reported to trigger β-catenin signaling in colon cancer, they display different oncogenic properties through distinct mechanisms, where EP2 enhances cancer cell growth through a Gs-axin-β-catenin signaling axis, and GPR48 promotes tumor metastasis through a PI3K-Akt-Erk-β-catenin-TCF signaling axis.4,5 These results are not surprising, because GPCRs have the ability to use alternative signaling pathways,47 and it is likely that these 2 GPCRs regulate distinct functional programs downstream of β-catenin through their respective signaling cascade in colon cancer. Furthermore, using clinically available Cox inhibitors, we and others have previously shown that inhibition of Cox suppresses β-catenin activation and has a profound effect on LSC functions in AML initiated by MLL-AF9 or AML1-ETO.1,6 However, it is not clear whether the Cox receptors EP1/EP2 also play an important role in AML leukemogenesis. Moreover, we cannot rule out the possibility that GPR108, GPR172b, and GPR177 identified in our microarray analysis have oncogenic potential through β-catenin signaling in MLL AML. In particular, it has been recently reported that GPR177 (also known as Wls) functions as a positive regulator of β-catenin signaling to affect cell proliferation in melanoma.48 The role of these GPCRs in leukemogenesis remains the subject of future studies.
In addition to the Wnt/β-catenin signaling, we show that GPR84 overexpression also activates an oncogenic transcription program including the induction of MLL-fusion target genes, which are known to be key components of the aberrant self-renewal–associated signature identified in MLL LSCs.17 Among those target genes, Meis1a and Hox genes are essential for the development of MLL.1,17,24,49 Meis1a expression is a necessary requirement to sustain proliferation and block differentiation in leukemic cells.50,51 Coexpression of Meis1a and Hoxa7 is a common occurrence in human AML and is sufficient to induce AML in mice.52-54 Eya1 and Six1 have also been found to be upregulated in human and mouse MLL AML.28 Eya1 has the capacity to immortalize hematopoietic stem/progenitor cells in vitro and cooperates with Six1 to potentiate its transforming capacity.28 Indeed, GPR84-mediated upregulation of these key MLL-fusion targets facilitates the enhanced survival and malignant transformation of pre-LSCs. This is consistent with our observation suggesting that GPR84 overexpression contributes to inducing murine full-blown stem cell–derived MLL, possibly through concurrent enhancement of a MLL transcriptional program and β-catenin signaling. Furthermore, our study demonstrates that GPR84 knockdown prevents leukemic cell dissemination to secondary organs, including the spleen. Interestingly, loss of Mef2c has been reported to compromise the invasiveness of MLL-ENL leukemic cells in vivo29 ; hence, it appears most likely that GPR84 knockdown–mediated downregulation of Mef2c is responsible for impaired dissemination of MLL leukemic cells during leukemia development. We also show that GPR84 deficiency inhibits clonogenic growth of MLL pre-LSCs and leads to a defect in cell-cycle progression at the G1-S phase transition. This can be attributed primarily to GPR84 knockdown–mediated upregulation of Ms4a3, which induces G1-phase cell-cycle arrest in HSCs and primary AML blasts.31,32,55 Collectively, our study therefore demonstrates a multifaceted regulatory function of GPR84 in leukemia formation and shows a strong dependence of MLL leukemic cells on GPR84 for growth and survival in vitro and in vivo. The dramatic effect of GPR84 on stem cell–derived MLL leukemogenesis suggests that GPR84 is a direct target of MLL-AF9–dependent transformation in vivo and highlights the importance of this signaling molecule in this disease.
In summary, our findings demonstrate a strong rationale for blocking the GPCR-β-catenin signaling axis as a therapeutic strategy to target stem cell–derived MLL. The potential therapeutic value of targeting GPCRs is also supported by the fact that GPCRs are highly tractable drug targets.9 Given that GPR84 expression can be significantly increased by a potent inflammation inducer in a human MLL AML cell line,16 it is likely that GPR84 promotes leukemogenesis through aberrant activation of inflammatory pathways in LSCs. Recent studies have shown that genes involved in the inflammatory response play a causative role in the development and maintenance of LSCs,56,57 implicating a link between inflammation and leukemia. Further studies will determine the precise molecular mechanisms of how proinflammatory receptor GPR84 regulates MLL-fusion target genes and β-catenin–dependent self-renewal programs that drive leukemic transformation. In the long term, it will be important to explore whether GPR84 and oncogenic GPCR pathways recently identified in AML (eg, inflammation-related Cox-EP1/EP2)1,6 share a common β-catenin and/or MLL target gene expression signature, and to investigate whether functional cross talk exists between different GPCR signaling pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Warren Alexander for helpful comments on the manuscript.
J.Y.W. is an Australian Research Council Future Fellow (FT120100612). P.A.D. is supported by the Steggles Scholarship. This work was supported by grants from the National Health and Medical Research Council (APP1045524) and the Cancer Institute New South Wales (11/CDF/3-38) to J.Y.W.
Authorship
Contribution: P.A.D. designed and performed experiments and analyzed data; C.Y. and E.G. performed western blot analysis; H.H.L.L. conducted microarray experiments and contributed to part of the mouse survival experiments; C.Y. and J.R.L performed real-time RT-PCR analysis; B.L. analyzed DNA microarray data; M.H., M.D.N., and J.W. provided reagents and reviewed the manuscript; and J.Y.W. designed experiments, supervised the project, and wrote the manuscript; and all authors reviewed and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jenny Y. Wang, Children’s Cancer Institute Australia, Lowy Cancer Research Centre, University of New South Wales, PO Box 81, Randwick, NSW 2031, Australia; e-mail: jenny.wang@unsw.edu.au.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal