Abstract
Basis.
Abnormalities in chromosome 8 (8p-/8q+) are observed in 2-5% of CLL patients. Microarray studies have revealed up to 30-40% of 8 alterations in del(17p) patients and an independent association with poor outcome. Large series assessing CLL patients with 8p-/8q+ are scarce.
Aims.
1. To describe the frequency of 8q gains (8q+) and 8p losses (8p-) in CLL patients with del(17p); 2. To compare cytogenetic and clinical characteristics between patients with 8p-/8q+ (Alt-chr8) and those with normal chromosome 8 (N-chr8); 3. To assess their prognostic value.
Patients and methods.
From 2,249 patients included in the Spanish CLL database, 75 del(17p) cases were selected. Gains of MYC (8q24) and losses of LPL (8p22) were studied by FISH. Clinical and cytogenetic data of Alt-chr8 and N-chr8 were compared.
Results.
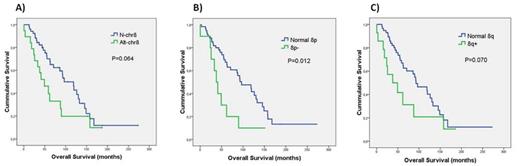
8p- and/or 8q+ were found in 21/75 patients (28%). In the Alt-chr8 group, 8q+ was more frequent than 8p- (71% vs. 52%) and 29% showed concomitance of both abnormalities, suggesting the presence of i(8q). Six different FISH patterns were identified, some of them coexisting in the same patient (Table 1). Conventional cytogenetics data were available in 47 cases (15 Alt-chr8 and 32 N-chr8). Alt-chr8 group showed a higher median number of alterations and frequency of complex karyotypes (P=0.048 and P=0.013). In the Alt-chr8 group, the karyotype revealed 8p-/8q+ in 3 patients and in 9 cases with abnormal karyotype, the presence of marker chromosomes, added material and/or cryptic alterations would explain the FISH results (Table 1). From 66 cases, routine FISH data (13q, 12 and 11q) were available and no significant differences were detected among Alt-chr8 and N-chr8, as with other clinical and analytical parameters at diagnosis. Of note, shorter Overall Survival (OS) was observed for Alt-chr8, although differences were only significant for patients with 8p- (P=0.012, Figure 1). Interestingly, for 3 patients of Alt-chr8 group, previous non-del(17p) samples already presented 8p-/8q+.
Conclusions.
1. In CLL patients with del(17p), detection of 8p- and/or 8q+ is associated with an increased karyotypic complexity and a worse outcome; 2. 8p-/8q+ could act as a primary event that trigger del(17p). More cases are required to confirm this hypothesis.
Acknowledgments.PI11/01621; RD12/0036/0044, RD12/0036/0069; 2014/SGR585; Fundació La Caixa.
Karyotypes and FISH results of patients with del(17p) and Alt-chr8.
| . | Conventional Cytogenetics . | FISH . | |||
|---|---|---|---|---|---|
| ID . | Karyotype . | % del(17p) . | Chromosome 8 alteration . | ||
| % . | Pattern* . | ||||
| 1 | 46,XX,del(8)(p21),add(10)(q26),add(17)(p13),+2ac[5]/47,XX,+12,add(17)(p13),del(18)(q21),add(22)(q13)[3] | 80 | 20 | 1O2G | 8p- |
| 2 | - | 95 | 75 | ||
| 3 | 46,XX,add(6)(q24),add(14)(q32,3),i(17)(q10)[6]/46,XX[8] | 95 | 75 | ||
| 4 | - | 76 | 50 | ||
| 5 | 45,XY,-5,-9,-15,add(17)(p13),+18,-21,+2mar[13]/46,XY[37] | 70 | 17 | ||
| 6 | 46,X,der(X),add(8)(p23),del(13)(q12q22),add(17)(p13)[11]/46,XX[13] | 10 | 32 | 1O3G | 8p- and 8q+ |
| 7 | - | 95 | 64 | ||
| 8 | 45,XY,add(3)(q29),del(4)(q26q35),der(7)(1p36-1p32::7p22-7q32::15q22-15q26), -8,der(9),del(13)(q21q34),-15,-17,-18,+19,add(19)(p13),+2mar,+ac[17]/46,XY[3] | 78 | 34/21 | 1O3G/2O3G | |
| 9 | 44,X,-X,-6,der(13;15)(q10;q10),add(17)(p13),-20,+mar[13]/46,XX[7] | 95 | 10/23 | 1O3G/1O2G | |
| 10 | - | 95 | 66/31 | ||
| 11 | 45,XY,add(6)(q22),del(11)(q11q22),-17[15]/44,XY,add(6)(q22),del(11)(q11q22),-17,-20,-22,+mar[2] | 87 | 40/24 | ||
| 12 | 46,XX,del(13)(q14q21)[2]/45,X,-X,del(13)(q14q21)[3]/45,XX,add(3)(q27), t(9;10)(q21;q22),+12,der(12)t(12;17)(q11;p11),del(13)(q14q21),-14,-17[7]/46,XX[8] | 70 | 62 | 2O3G | 8q+ |
| 13 | 46,XY[30] | 14 | 88 | ||
| 14 | 46,XY[13] | 80 | 82 | ||
| 15 | 47,XY,+12[8]/46,XY,add(1)(p34),add(2)(q34),t(11;22)(p14;q11),+12,-22[15]/46,XY[11] | 75 | 18 | ||
| 16 | 43,X,-X,del(2)(p15),+4,-7,add(11)(q21),-12,-13,add(14)(q32),add(17)(p11)[6]/46,XX[9] | 19 | 14 | ||
| 17 | 45,XY,del(6)(q?),-9,add(14)(q32),-22,+mar[9]/ 46,XY,del(6)(q?),add(17)(p13),add(19)(q13)[21] | 55 | 23 | ||
| 18 | 45,XY,add(6)(p11),-22[13]/46,XY,i(17)(q10)[5]/46,XY[16] | 16 | 57 | ||
| 19 | - | 70 | 66 | 2O4G | |
| 20 | - | 90 | 81 | 2OnG | |
| 21 | 46,XY[11] | 43 | 60 | 4O4G | Tetraploid |
| . | Conventional Cytogenetics . | FISH . | |||
|---|---|---|---|---|---|
| ID . | Karyotype . | % del(17p) . | Chromosome 8 alteration . | ||
| % . | Pattern* . | ||||
| 1 | 46,XX,del(8)(p21),add(10)(q26),add(17)(p13),+2ac[5]/47,XX,+12,add(17)(p13),del(18)(q21),add(22)(q13)[3] | 80 | 20 | 1O2G | 8p- |
| 2 | - | 95 | 75 | ||
| 3 | 46,XX,add(6)(q24),add(14)(q32,3),i(17)(q10)[6]/46,XX[8] | 95 | 75 | ||
| 4 | - | 76 | 50 | ||
| 5 | 45,XY,-5,-9,-15,add(17)(p13),+18,-21,+2mar[13]/46,XY[37] | 70 | 17 | ||
| 6 | 46,X,der(X),add(8)(p23),del(13)(q12q22),add(17)(p13)[11]/46,XX[13] | 10 | 32 | 1O3G | 8p- and 8q+ |
| 7 | - | 95 | 64 | ||
| 8 | 45,XY,add(3)(q29),del(4)(q26q35),der(7)(1p36-1p32::7p22-7q32::15q22-15q26), -8,der(9),del(13)(q21q34),-15,-17,-18,+19,add(19)(p13),+2mar,+ac[17]/46,XY[3] | 78 | 34/21 | 1O3G/2O3G | |
| 9 | 44,X,-X,-6,der(13;15)(q10;q10),add(17)(p13),-20,+mar[13]/46,XX[7] | 95 | 10/23 | 1O3G/1O2G | |
| 10 | - | 95 | 66/31 | ||
| 11 | 45,XY,add(6)(q22),del(11)(q11q22),-17[15]/44,XY,add(6)(q22),del(11)(q11q22),-17,-20,-22,+mar[2] | 87 | 40/24 | ||
| 12 | 46,XX,del(13)(q14q21)[2]/45,X,-X,del(13)(q14q21)[3]/45,XX,add(3)(q27), t(9;10)(q21;q22),+12,der(12)t(12;17)(q11;p11),del(13)(q14q21),-14,-17[7]/46,XX[8] | 70 | 62 | 2O3G | 8q+ |
| 13 | 46,XY[30] | 14 | 88 | ||
| 14 | 46,XY[13] | 80 | 82 | ||
| 15 | 47,XY,+12[8]/46,XY,add(1)(p34),add(2)(q34),t(11;22)(p14;q11),+12,-22[15]/46,XY[11] | 75 | 18 | ||
| 16 | 43,X,-X,del(2)(p15),+4,-7,add(11)(q21),-12,-13,add(14)(q32),add(17)(p11)[6]/46,XX[9] | 19 | 14 | ||
| 17 | 45,XY,del(6)(q?),-9,add(14)(q32),-22,+mar[9]/ 46,XY,del(6)(q?),add(17)(p13),add(19)(q13)[21] | 55 | 23 | ||
| 18 | 45,XY,add(6)(p11),-22[13]/46,XY,i(17)(q10)[5]/46,XY[16] | 16 | 57 | ||
| 19 | - | 70 | 66 | 2O4G | |
| 20 | - | 90 | 81 | 2OnG | |
| 21 | 46,XY[11] | 43 | 60 | 4O4G | Tetraploid |
*O: LPL signal in orange, G: MYC signal in green.
Kaplan Meier plots for OS and (A) 8p- and/or 8q+, (B) 8p- or (C) 8q+.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal