Abstract
Background: The myelodysplastic syndromes (MDS) are a heterogeneous group of malignancies characterized by multiple genomic abnormalities. New technologies are needed that bridge the complex molecular biology of MDS and available therapeutics.
Methods: Therefore, we developed a predictive simulation software program to create MDS patient avatars based on disease-specific genomic profile. The avatar technology has three layers: the first layer is a proprietary mathematics solver engine architected to handle millions of differential equations representing biological cross-talk reactions needed for modeling MDS physiology; the middle layer is the comprehensive functional proteomics representation of disease physiology network; and the topmost layer is a semiconductor engineering-based automaton engine which allows high-throughput simulation of multi-million interventions through assembly code language. The MDS avatars were intended to map the complex interplay of dysregulated pathways and predict potential therapeutics. The human MDS-L cell line is one of few bona fide MDS cell lines and was analyzed by conventional G-banding karyotyping, FISH, array CGH and Sanger sequencing of MDS-relevant genes. Using this genomic information, a simulation patient avatar was created. Next, a library of over 80 FDA approved agents was simulated against the dysregulated pathways. A list of predicted drugs where then prospectively validated using in vitro culture of MDS-L cells.
Results: The MDS-L cells harbored an activating NRAS mutation (c.35G>C, p.G12A) and complex chromosome abnormalities. The simulation MDS-L patient avatar predicted activation of the RAF-ERK pathway due to NRAS mutation and various copy number variations (CNVs) including high CN of IGFR, AURKA, PAR5 that predicted high AKT activation, high CN of mTOR, high CN of IL6 and JAK3 with predicted activation of STAT3 and STAT5, high CN of MDM4 with predicted lower levels of TP53, high CN of RCE1, MAPK1 and MAP2K1, and low CN of RASA1 and DUSP1 (Figure 1). Given the strong dominance of AKT and ERK loops in this profile (Figure 1), we simulated potential drugs individually and in combinations to predict impact on these pathways. Iterative simulations identified four FDA-approved drugs: (A) nelfinavir and celecoxib acting on AKT, (B) sorafenib and trametinib that inhibit RAF-ERK targets, and (C) statins that inhibit the RAS-RAF-ERK pathway through inhibition of prenylation. In vitro validation experiments showed statistically significant reductions in MDS-L cell viability when treating with nelfinavir alone, celecoxib alone, and enhanced additive reduction in viability with the combination of nelfinavir and celecoxib (Table 1).
Conclusions: This study shows how a novel simulation method can be employed to use patient-specific MDS genomic profiling to map dysregulated pathways and predict potentially therapeutic agents. The MDS case presented led to the discovery of a novel combination of nelfinavir and celecoxib. Results from this study serve as the basis for MDS clinical trials that assign treatment based on genetic mutations.
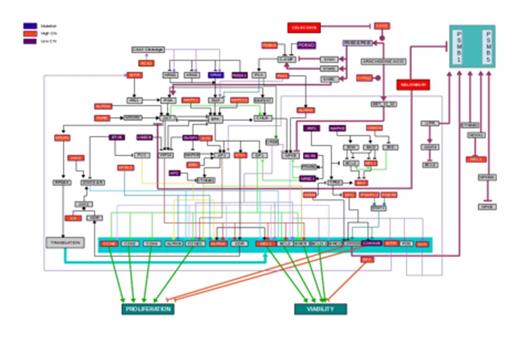
Predicted Dysregulated Pathways in MDS-L Cells. MDS-L genomic data was used to generate a map of dysregulated pathways. NRAS mutation is highlighted in blue with mapping of consequent downstream effects. High Copy Number (CN) in red and Low (CN) mutations in purple are also mapped with predicted downstream pathway effects. The totality of dysregulated pathways is predicted to converge on increased cell proliferation and increased cell viability. FDA-approved drugs predicted to impact dysregulated pathways (i.e., celecoxib and nelfinavir) are also shown in the map at critical impact points.
Predicted Dysregulated Pathways in MDS-L Cells. MDS-L genomic data was used to generate a map of dysregulated pathways. NRAS mutation is highlighted in blue with mapping of consequent downstream effects. High Copy Number (CN) in red and Low (CN) mutations in purple are also mapped with predicted downstream pathway effects. The totality of dysregulated pathways is predicted to converge on increased cell proliferation and increased cell viability. FDA-approved drugs predicted to impact dysregulated pathways (i.e., celecoxib and nelfinavir) are also shown in the map at critical impact points.
MDS-L Cell In Vitro Validation Experiments of Drug Treatments.
| . | . | Nelfinavir . | ||
|---|---|---|---|---|
| Drug Dose | 0μM | 5μM | 10μM | |
| Celecoxib | 0 μM | 100.00% | 81.45% ± 7.8% | 53.49% ± 6.9% |
| 30 μM | 71.34% ± 4.89% | 60.58% ± 3.25% | 35.78% ± 3.59% | |
| . | . | Nelfinavir . | ||
|---|---|---|---|---|
| Drug Dose | 0μM | 5μM | 10μM | |
| Celecoxib | 0 μM | 100.00% | 81.45% ± 7.8% | 53.49% ± 6.9% |
| 30 μM | 71.34% ± 4.89% | 60.58% ± 3.25% | 35.78% ± 3.59% | |
MDS-L cells were treated with vehicle control, nelfinavir alone (incremental dose increases), celecoxib alone (incremental dose increases), and combination nelfinavir and celecoxib. MDS-L cell viability was quantified and compared among treatment groups. Both nelfinavir alone and celecoxib alone reduce MDS-L cell viability in a dose-dependent manner. The combination of nelfinavir and celecoxib showed enhanced additive reduction of MDS-L cell viability, again in a dose-dependent manner.
Cogle:PHAR: Consultancy. Vali:CellWorks: Employment. Kumar:CellWorks: Employment. Singh:CellWorks: Employment. Tyagi:CellWorks: Employment. Abbasi:CellWorks: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal