Abstract
Myelodysplastic syndromes (MDS) are a group of chronic and often progressive myeloid neoplasms associated with remarkable heterogeneity in the degree of cytopenia, morphology and clinical course. Various somatic mutations are involved in the pathogenesis of MDS. The recent identification of acquired mutations in the key components of the spliceosome machinery strongly implicates abnormalities of mRNA splicing in the pathogenesis of myelodysplastic syndromes.
In this study, we performed Targeted Massively Parallel Sequencing on a custom 29 myeloid gene panel (all coding regions) of myeloid genes using an AmpliSeq approach (Life Tech) on 148 MDS patient samples. Barcodes were added using NEB kit and libraries were prepared for Illumina HiSeq2500 sequencing.
Results: The median age of patients was 72 (19-97) years and 65% patients were male. Majority of patients were RA-RS, RCMD, RCMD-RS, RAEB1 and RAEB2. Conventional bone marrow cytogenetic was normal in 80/148 (55%) of cases while sequencing of 29-gene panel revealed at least one mutation in 131/148 (88%) patients. Importantly a combination of cytogenetic and mutation screening detected genetic abnormalities in 141/148 (95%) of MDS patients, thus highlighting the importance of combining targeted mutation screening along with conventional karyotyping in MDS patients.
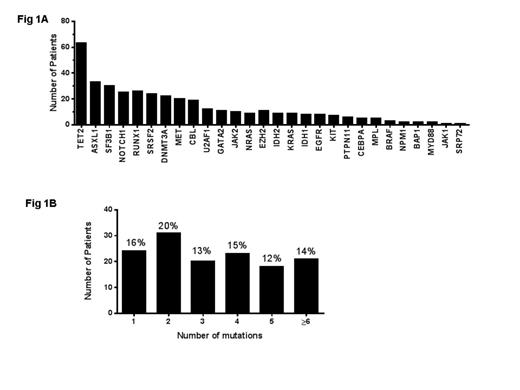
The majority of patients had more ≥2 mutations 112/148 (75%) suggesting clonal heterogeneity. As shown in Figure (1A), the most frequently mutated genes were ASXL1, SF3B1, NOTCH1, TET2, SRSF2, RUNX1 and DNMT3A. Mutations in the spliceoosome complex genes were detected in 66/148 (44%) of patients with SF3B1 (30/148; 20%) more frequent than SRSF2 (24/148; 16%) and U2AF1 (12/148; 8%). These mutations were mutually exclusive except for one patient.
SF3B1 mutations were detected in 95% (19/20) of RA-RS and RCMD-RS cases, while >50% of patients with SRSF2 mutations had RAEB1 or RAEB2. Mutations in the spliceosome complex co-occur with mutations in the epigenetic modifiers such as DNMT3A and TET2 mutations which is consistent with the previous report.
We identified 47 patients with 2 (n=37) or three (n=10) TET2 mutations. Of these, 16 patients had mutations at significantly different allele frequencies suggesting that independent clones had arisen, raising the possibility that in these samples a pre-leukemic genetic or epigenetic cellular environment exists that selects for TET2 mutations. We also saw a similar trend, although to a lesser extent, for NOTCH1, RUNX1 and NRAS mutations.
Conclusions: Combination of targeted sequencing of most commonly mutated genes and conventional cytogenetic can detect genetic abnormalities in 95% of MDS cases. This study shows that >75% of MDS cases have more than one mutations demonstrating clonal heterogeneity. SF3B1 mutations are detected in 95% of RA-RS and RCMD-RS cases and are frequently associated with mutations in epigenetic modifiers. A high level of mutation detection has been possible because detection of low level mutations due to deep sequencing.
(A) The most frequently mutated genes were TET2, ASXL1, SF3B1, SRSF2, DNMT3A and RUNX1 (B) Majority of patients had two or more mutations.
(A) The most frequently mutated genes were TET2, ASXL1, SF3B1, SRSF2, DNMT3A and RUNX1 (B) Majority of patients had two or more mutations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal