Abstract
Background: In a subset of CML patients treated with tyrosine kinase inhibitors (TKI) primary or secondary resistance has been observed. Depending on disease state and treatment, in around 30-90% of these cases secondary mutations in the ABL1 part of the BCR-ABL1 fusion gene have been described which became clear indicators for change of therapy. In addition, also cytogenetic evolution is correlated to TKI resistance. However, in a high percentage of cases the reason for TKI resistance remains unclear, yet.
Aim: We hypothesized that mutations in genes frequently involved in myeloid malignancies may contribute to resistance to TKI.
Methods: We investigated 34 CML patients with no response or loss of response to TKI defined by clinical parameters and a BCR-ABL1 level >10%. Median age of patients was 54 years (range: 33-88 years), 22 were male and 12 female. At the time point of analysis 9 patients had received imatinib only, 16 patients had received 2 TKIs, 7 had 3 TKIs, and 2 even 4 TKIs (one of those in addition had undergone stem cell transplantation, SCT). In 24 cases primary resistance and in 10 cases secondary resistance after initial response to imatinib was observed. Samples for subsequent analyses were from the time point of BCR-ABL1 mutation analysis performed upon clinically suspected TKI resistance and in none any BCR-ABL1 mutation was identified by Sanger sequencing at a sensitivity level of 10-20%. For these samples we applied a pan-myeloid panel of 32 genes: ASXL1, BCOR, BRAF, CALR, CBL, CSF3R, DNMT3A, ETV6, EZH2, FLT3-TKD, GATA1, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PHF6, PTPN11, SETBP1, SF1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, ZRSR2. Either complete coding regions or hotspots were first amplified by a microdroplet-based assay (RainDance, Billerica, MA) and subsequently sequenced with a MiSeq instrument (Illumina, San Diego, CA). In addition, RUNX1 was sequenced on the 454 NGS platform (454 Life Sciences, Branford, CT). For backtracking of mutations also deep sequencing by 454 technology was performed.
Results: In 18 of the 34 patients (52.9%) mutations were detected. In a large subset of 12 cases (66.7%) ASXL1 was the only mutated gene. The other 6 cases had mutations in 1) RUNX1, 2) IDH1, 3) DNMT3A, 4) BCOR, 5) DNMT3A and WT1, and 6) NRAS, PTPN11, RUNX1 and WT1, respectively. Median mutation loads were 25% (range 4-53%). In order to assess the kinetics and onset of these mutations backtracking in 91 prior samples was performed.
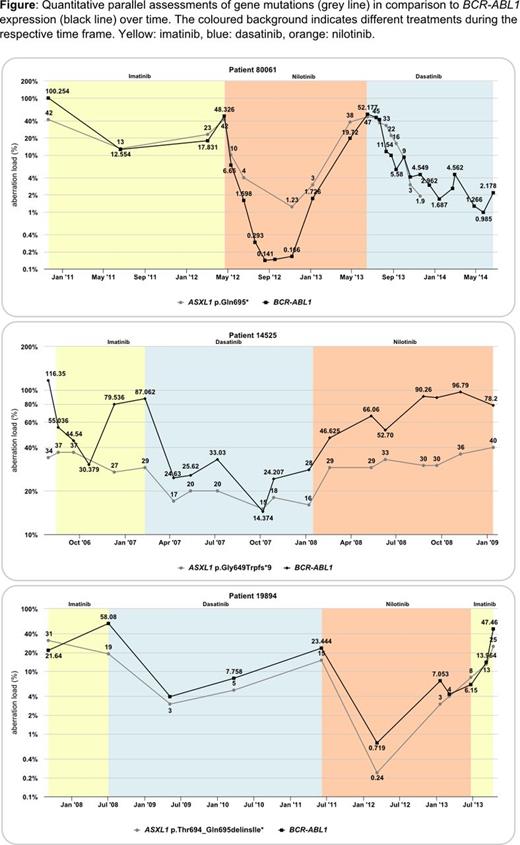
11 of the 12 ASXL1 mutations were detected in cases with primary TKI resistance. In 7 of these patients samples from diagnosis of CML were available and ASXL1 mutations were already present at high levels in all of them. The pattern obtained from the backtracking analysis clearly showed that the ASXL1 mutation load closely correlates with the BCR-ABL1 expression (3 examples are shown in the figure). In only one patient with secondary resistance the ASXL1 mutation was gained after 40 months of imatinib treatment. Results of the other 6 patients are as listed in the following (numbering as indicated above). 1) A RUNX1 mutation with 40% load was detected after 6 months of dasatinib treatment. This case was negative for this mutation before and during 2 years of imatinib treatment. 2) An IDH1 mutation was gained 1 year after start of nilotinib treatment. 3) and 4) Backtracking is ongoing in patients. 5) DNMT3A and WT1 mutations both were gained after 34 months of imatinib treatment. 6) The 4 mutations in this patient were not present at diagnosis and detected for the first time after 22 months of treatment with 3 consecutive TKIs and upon non-response to allogeneic SCT. These results show that mutations can occur during any kind of TKI treatment, but also are frequently present already at the time of CML diagnosis. These gene mutations are considered to contribute to TKI resistance in both scenarios.
Conclusions: 1) Mutations known from other myeloid malignancies are frequently present in CML patients that do not respond to or lost response to TKI treatment and lack BCR-ABL1 resistance mutations. 2) The courses of these mutations clearly parallelize with the courses of the BCR-ABL1 positive clones. 3) ASXL1 is the most frequently mutated gene. 4) These mutations are additional genetic hits that point to a more aggressive and treatment resistant biology of the BCR-ABL1 positive clone and 5) may indicate that alternative therapies besides TKI inhibitors are needed in these cases.
Schnittger:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Nadarajah:MLL Munich Leukemia Laboratory: Employment. Meggendorfer:MLL Munich Leukemia Laboratory: Employment. Fasan:MLL Munich Leukemia Laboratory: Employment. Rose:MLL Munich Leukemia Laboratory: Employment. Alpermann:MLL Munich Leukemia Laboratory: Employment. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal