Abstract
Acute myeloid leukemia (AML) is characterized by developmental arrest which is thought to arise from transcriptional dysregulation of myeloid development programs. Transcription factor (TF) dysregulation has been identified at both the genomic and transcriptomic levels; however the actual transcriptional environment in AML patients characterized by the relative abundance of TF protein expression compared with normal blasts has not yet been described. Here, we have analysed the nuclear proteome of AML blasts in comparison with normal CD34+ cells and carried out a parallel analysis of the transcriptome using Affymetrix arrays.
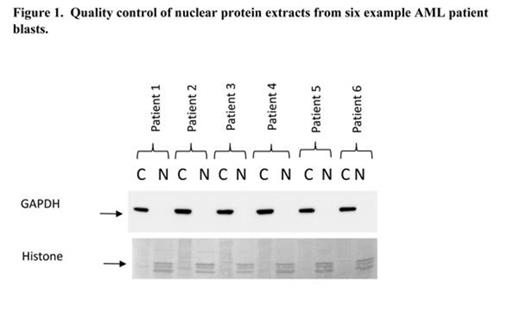
In our experimental design we restricted our analysis to the minimally differentiated FAB M1 since this subtype has little developmental heterogeneity and would also be developmentally matched to normal controls. The final experimental design comprised nuclear protein extracts from 5 normal CD34+ controls and 15 FAB M1 patients (>80% viable blasts; <10% CD14+/CD15+ cells). Purity of nuclear fractions was assessed by western blotting for histone and GAPDH (Figure 1). Nuclear tryptic peptides were generated and in each experiment labelled with 8 channel isobaric tagging to allow relative quantification coupled with peptide/protein identification using tandem mass spectrometry. In parallel mRNA from these samples were analysed using Human Transcriptome Array 2.0 (Affymetrix, USA).
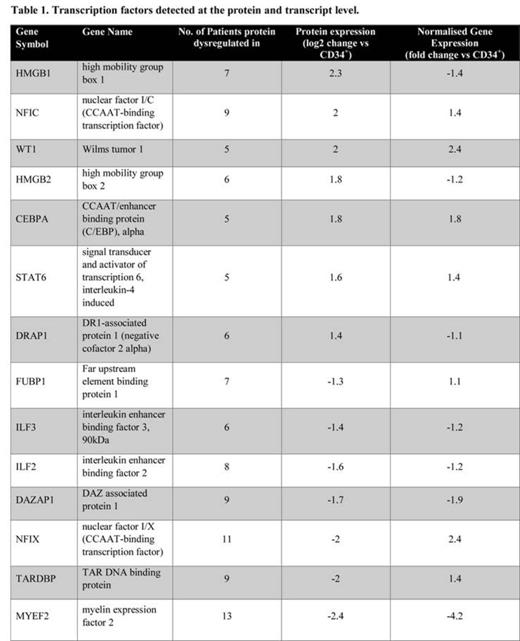
In total 6164 proteins were found and quantified. After applying appropriate quality control criteria 437 proteins were found to be significantly dysregulated between normal CD34+ cells and AML blasts. To identify frequently dysregulated proteins we selected those that consistently changed in at least 5 AML patients (± >1.2 fold). This yielded 188 proteins of which 112 (60%) were nuclear proteins. We identified 7 upregulated transcription factors in leukaemic cells compared to normal CD34+ cells; 3 of which had been previously associated with AML (CEBPA, STAT6 and WT1). Comparative analysis of mRNA of these changes showed that these increases were also significantly observed at the transcriptional level (Table 1). The remainder (DRAP1, NFIC, HMGB1 and HMGB2) had not been previously reported in AML, and none of the changes were seen at the transcriptional level indicating increased expression arose at post-transcriptionally or were due to an increased level of nuclear localization. We also identified 7 down-regulated TF, one of which had been previously associated with AML (DAZAP1) with the remaining being newly described abnormalities (MYEF2, NFIX, FUBP1, TARDBP, ILF2, ILF3). Again most of these changes (4 of 6) were not seen at the transcriptional level. We also observed changes in 15 heterogeneous nuclear ribonucleoproteins (hnRNP) affecting mRNA processing (including: A0, A1, A2B1, A3, AB, C, D, DL, PF, H1, H3, K, L, R and UL2) and we are currently examining whether their expression correlates with increased alternative splicing that we have observed in these patients from analysis of the exon arrays.
These data are the first analysis of the nuclear proteome in AML and have identified changes in transcription factor expression that would not have been seen at the mRNA level. We are performing in silico analysis to determine whether dysregulation of these TF give rise to corresponding changes in known target genes.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal